LGR5 expressing skin fibroblasts define a major cellular hub perturbed in scleroderma
- PMID: 35381199
- PMCID: PMC7612792
- DOI: 10.1016/j.cell.2022.03.011
LGR5 expressing skin fibroblasts define a major cellular hub perturbed in scleroderma
Abstract
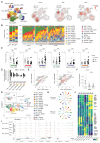
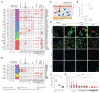
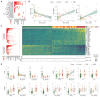
Systemic sclerosis (scleroderma, SSc) is an incurable autoimmune disease with high morbidity and mortality rates. Here, we conducted a population-scale single-cell genomic analysis of skin and blood samples of 56 healthy controls and 97 SSc patients at different stages of the disease. We found immune compartment dysfunction only in a specific subtype of diffuse SSc patients but global dysregulation of the stromal compartment, particularly in a previously undefined subset of LGR5+-scleroderma-associated fibroblasts (ScAFs). ScAFs are perturbed morphologically and molecularly in SSc patients. Single-cell multiome profiling of stromal cells revealed ScAF-specific markers, pathways, regulatory elements, and transcription factors underlining disease development. Systematic analysis of these molecular features with clinical metadata associates specific ScAF targets with disease pathogenesis and SSc clinical traits. Our high-resolution atlas of the sclerodermatous skin spectrum will enable a paradigm shift in the understanding of SSc disease and facilitate the development of biomarkers and therapeutic strategies.
Keywords: FOS; GVHD; LGR5; TWIST1; autoimmune; fibroblast; fibrosis; skin; systemic sclerosis.
Copyright © 2022 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures





References
-
- Benyamine A, Magalon J, Sabatier F, Lyonnet L, Robert S, Dumoulin C, Morange S, Mazodier K, Kaplanski G, Reynaud-Gaubert M, et al. Natural killer cells exhibit a peculiar phenotypic profile in systemic sclerosis and are potent inducers of endothelial microparticles release. Front Immunol. 2018;9:1665. doi: 10.3389/FIMMU.2018.01665. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

