The genetic architecture of flowering time changes in pea from wild to crop
- PMID: 35383838
- PMCID: PMC9238443
- DOI: 10.1093/jxb/erac132
The genetic architecture of flowering time changes in pea from wild to crop
Abstract
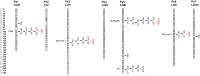
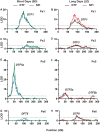
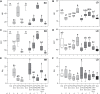
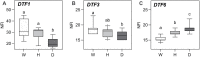
Change in phenology has been an important component in crop evolution, and selection for earlier flowering through a reduction in environmental sensitivity has helped broaden adaptation in many species. Natural variation for flowering in domesticated pea (Pisum sativum L.) has been noted and studied for decades, but there has been no clear account of change relative to its wild progenitor. Here we examined the genetic control of differences in flowering time between wild P. sativum ssp. humile and a typical late-flowering photoperiodic P. s. sativum accession in a recombinant inbred population under long and short photoperiods. Our results confirm the importance of the major photoperiod sensitivity locus Hr/PsELF3a and identify two other loci on chromosomes 1 (DTF1) and 3 (DTF3) that contribute to earlier flowering in the domesticated line under both photoperiods. The domesticated allele at a fourth locus on chromosome 6 (DTF6) delays flowering under long days only. Map positions, inheritance patterns, and expression analyses in near-isogenic comparisons imply that DTF1, DTF3, and DTF6 represent gain-of-function alleles of the florigen/antiflorigen genes FTa3, FTa1, and TFL1c/LF, respectively. This echoes similar variation in chickpea and lentil, and suggests a conserved route to reduced photoperiod sensitivity and early phenology in temperate pulses.
Keywords: FT genes; Pisum; Adaptation; QTL analysis; florigen; flowering time; genetics; legume; pea; phenology; photoperiod.
© The Author(s) 2022. Published by Oxford University Press on behalf of the Society for Experimental Biology. All rights reserved. For permissions, please email: journals.permissions@oup.com.
Figures

Comment in
-
Putting the pea in photoPEAriod.J Exp Bot. 2022 Jun 24;73(12):3825-3827. doi: 10.1093/jxb/erac170. J Exp Bot. 2022. PMID: 35749691 Free PMC article.
References
-
- Ben-Ze’ev N, Zohary D.. 1973. Species relationships in the genus Pisum. Israel Journal of Botany 22, 73–91.
-
- Bett K, Ramsay L, Chan C, et al. . 2016. Lentil 1.0 and beyond. In: Plant and Animal Genomes XXIV Conference, San Diego, CA, 9–13.
-
- Broman KW, Wu H, Sen S, Churchill GA.. 2003. R/qtl: QTL mapping in experimental crosses. Bioinformatics 19, 889–890. - PubMed
-
- Cockram J, Jones H, Leigh FJ, O’Sullivan D, Powell W, Laurie DA, Greenland AJ.. 2007. Control of flowering time in temperate cereals: genes, domestication, and sustainable productivity. Journal of Experimental Botany 58, 1231–1244. - PubMed
-
- Doyle JJ, Doyle JL.. 1990. Isolation of plant DNA from fresh tissue. Focus 12, 13–15.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

