The Age of Next-Generation Therapeutic-Microbe Discovery: Exploiting Microbe-Microbe and Host-Microbe Interactions for Disease Prevention
- PMID: 35384688
- PMCID: PMC9119102
- DOI: 10.1128/iai.00589-21
The Age of Next-Generation Therapeutic-Microbe Discovery: Exploiting Microbe-Microbe and Host-Microbe Interactions for Disease Prevention
Abstract
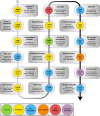
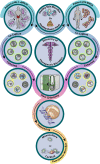
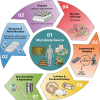
Humans are considered "superorganisms," harboring a diverse microbial collective that outnumbers human cells 10 to 1. Complex and gravely understudied host- and microbe-microbe interactions-the product of millions of years of host-microbe coevolution-govern the superorganism in almost every aspect of life functions and overall well-being. Abruptly disrupting these interactions via extrinsic factors has undesirable consequences for the host. On the other hand, supplementing commensal or beneficial microbes may mitigate perturbed interactions or enhance the interactive relationships that ultimately benefit all parties. Hence, immense efforts have focused on dissecting the innumerable host- and microbe-microbe relationships to characterize if a "positive" or "negative" interaction is at play and to exploit such behavior for broader implications. For example, microbiome research has worked to identify and isolate naturally antipathogenic microbes that may offer therapeutic potential either in a direct, one-on-one application or by leveraging its unique metabolic properties. However, the discovery and isolation of such desired therapeutic microbes from complex microbiota have proven challenging. Currently, there is no conventional technique to universally and functionally screen for these microbes. With this said, we first describe in this review the historical (probiotics) and current (fecal microbiota or defined consortia) perspectives on therapeutic microbes, present the discoveries of therapeutic microbes through exploiting microbe-microbe and host-microbe interactions, and detail our team's efforts in discovering therapeutic microbes via our novel microbiome screening platform. We conclude this minireview by briefly discussing challenges and possible solutions with therapeutic microbes' applications and paths ahead for discovery.
Keywords: defined microbial consortia; fecal microbiota transplant; host-microbe interactions; microbe-microbe interactions; next-generation sequencing; probiotics; therapeutic microbes; unculturables.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Therapeutic targeting of the host-microbiota-immune axis: implications for precision health.Front Immunol. 2025 Apr 29;16:1570233. doi: 10.3389/fimmu.2025.1570233. eCollection 2025. Front Immunol. 2025. PMID: 40364844 Free PMC article. Review.
-
Shaping the leaf microbiota: plant-microbe-microbe interactions.J Exp Bot. 2021 Jan 20;72(1):36-56. doi: 10.1093/jxb/eraa417. J Exp Bot. 2021. PMID: 32910810 Free PMC article.
-
Feed, Microbiota, and Gut Immunity: Using the Zebrafish Model to Understand Fish Health.Front Immunol. 2020 Feb 5;11:114. doi: 10.3389/fimmu.2020.00114. eCollection 2020. Front Immunol. 2020. PMID: 32117265 Free PMC article. Review.
-
Exploring the Human Microbiome: The Potential Future Role of Next-Generation Sequencing in Disease Diagnosis and Treatment.Front Immunol. 2019 Jan 7;9:2868. doi: 10.3389/fimmu.2018.02868. eCollection 2018. Front Immunol. 2019. PMID: 30666248 Free PMC article. Review.
-
Microbial Hub Taxa Link Host and Abiotic Factors to Plant Microbiome Variation.PLoS Biol. 2016 Jan 20;14(1):e1002352. doi: 10.1371/journal.pbio.1002352. eCollection 2016 Jan. PLoS Biol. 2016. PMID: 26788878 Free PMC article.
Cited by
-
New approaches to secondary metabolite discovery from anaerobic gut microbes.Appl Microbiol Biotechnol. 2025 Jan 20;109(1):12. doi: 10.1007/s00253-024-13393-y. Appl Microbiol Biotechnol. 2025. PMID: 39831966 Free PMC article. Review.
-
Photo-addressable microwell devices for rapid functional screening and isolation of pathogen inhibitors from bacterial strain libraries.Biomicrofluidics. 2024 Feb 29;18(1):014107. doi: 10.1063/5.0188270. eCollection 2024 Jan. Biomicrofluidics. 2024. PMID: 38434239 Free PMC article.
-
A Comprehensive Overview of Postbiotics with a Special Focus on Discovery Techniques and Clinical Applications.Foods. 2024 Sep 17;13(18):2937. doi: 10.3390/foods13182937. Foods. 2024. PMID: 39335866 Free PMC article. Review.
-
Working together: gut microbe-microbe interactions shape host inflammation.Infect Immun. 2025 Jul 8;93(7):e0051224. doi: 10.1128/iai.00512-24. Epub 2025 Jun 13. Infect Immun. 2025. PMID: 40512008 Free PMC article. Review.
-
The porcine skin microbiome exhibits broad fungal antagonism.Fungal Genet Biol. 2024 Aug;173:103898. doi: 10.1016/j.fgb.2024.103898. Epub 2024 May 28. Fungal Genet Biol. 2024. PMID: 38815692 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

