Silencing the conserved small nuclear ribonucleoprotein SmD1 target gene alters susceptibility to root-knot nematodes in plants
- PMID: 35385078
- PMCID: PMC9237699
- DOI: 10.1093/plphys/kiac155
Silencing the conserved small nuclear ribonucleoprotein SmD1 target gene alters susceptibility to root-knot nematodes in plants
Abstract
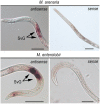
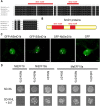
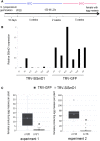
Root-knot nematodes (RKNs) are among the most damaging pests of agricultural crops. Meloidogyne is an extremely polyphagous genus of nematodes that can infect thousands of plant species. A few genes for resistance (R-genes) to RKN suitable for use in crop breeding have been identified, but virulent strains and species of RKN have emerged that render these R-genes ineffective. Secretion of RKN effectors targeting plant functions mediates the reprogramming of root cells into specialized feeding cells, the giant cells, essential for RKN development and reproduction. Conserved targets among plant species define the more relevant strategies for controlling nematode infection. The EFFECTOR18 (EFF18) protein from M. incognita interacts with the spliceosomal small nuclear ribonucleoprotein D1 (SmD1) in Arabidopsis (Arabidopsis thaliana), disrupting its function in alternative splicing regulation and modulating the giant cell transcriptome. We show here that EFF18 is a conserved RKN-specific effector that targets this conserved spliceosomal SmD1 protein in Solanaceae. This interaction modulates alternative splicing events produced by tomato (Solanum lycopersicum) in response to M. incognita infection. The alteration of SmD1 expression by virus-induced gene silencing in Solanaceae affects giant cell formation and nematode development. Thus, our work defines a promising conserved SmD1 target gene to develop broad resistance for the control of Meloidogyne spp. in plants.
© American Society of Plant Biologists 2022. All rights reserved. For permissions, please email: journals.permissions@oup.com.
Figures




References
-
- de Almeida Engler J, Gheysen G (2013) Nematode-induced endoreduplication in plant host cells: why and how? Mol Plant-Microbe Interact 26: 17–24 - PubMed
-
- Blanc-Mathieu R, Perfus-Barbeoch L, Aury J-MM, Da Rocha M, Gouzy J, Sallet E, Martin-Jimenez C, Bailly-Bechet M, Castagnone-Sereno P, Flot J-F, et al. (2017) Hybridization and polyploidy enable genomic plasticity without sex in the most devastating plant-parasitic nematodes. PLoS Genet 13: e1006777. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

