Retinitis pigmentosa-linked mutation in DHX38 modulates its splicing activity
- PMID: 35385551
- PMCID: PMC8985939
- DOI: 10.1371/journal.pone.0265742
Retinitis pigmentosa-linked mutation in DHX38 modulates its splicing activity
Erratum in
-
Correction: Retinitis pigmentosa-linked mutation in DHX38 modulates its splicing activity.PLoS One. 2023 Jan 20;18(1):e0280904. doi: 10.1371/journal.pone.0280904. eCollection 2023. PLoS One. 2023. PMID: 36662708 Free PMC article.
Abstract
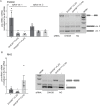
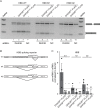
Retinitis pigmentosa (RP) is a hereditary disease affecting tens of thousands of people world-wide. Here we analyzed the effect of an amino acid substitution in the RNA helicase DHX38 (Prp16) causing RP. DHX38 has been proposed as the helicase important for the 2nd step of splicing. We showed that DHX38 associates with key splicing factors involved in both splicing steps but did not find any evidence that the RP mutations changes DHX38 interaction profile with the spliceosome. We further downregulated DHX38 and monitored changes in splicing. We observed only minor perturbations of general splicing but detected modulation of ~70 alternative splicing events. Next, we probed DHX38 function in splicing of retina specific genes and found that FSCN2 splicing is dependent on DHX38. In addition, RHO splicing was inhibited specifically by expression of DHX38 RP variant. Finally, we showed that overexpression of DHX38 promotes usage of canonical as well as cryptic 5' splice sites in HBB splicing reporter. Together, our data show that DHX38 is a splicing factor that promotes splicing of cryptic splice sites and regulate alternative splicing. We further provide evidence that the RP-linked substitution G332D modulates DHX38 splicing activity.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

