Structure-Based Evolution of G Protein-Biased μ-Opioid Receptor Agonists
- PMID: 35385593
- PMCID: PMC9322534
- DOI: 10.1002/anie.202200269
Structure-Based Evolution of G Protein-Biased μ-Opioid Receptor Agonists
Abstract
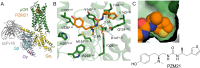
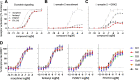
The μ-opioid receptor (μOR) is the major target for opioid analgesics. Activation of μOR initiates signaling through G protein pathways as well as through β-arrestin recruitment. μOR agonists that are biased towards G protein signaling pathways demonstrate diminished side effects. PZM21, discovered by computational docking, is a G protein biased μOR agonist. Here we report the cryoEM structure of PZM21 bound μOR in complex with Gi protein. Structure-based evolution led to multiple PZM21 analogs with more pronounced Gi protein bias and increased lipophilicity to improve CNS penetration. Among them, FH210 shows extremely low potency and efficacy for arrestin recruitment. We further determined the cryoEM structure of FH210 bound to μOR in complex with Gi protein and confirmed its expected binding pose. The structural and pharmacological studies reveal a potential mechanism to reduce β-arrestin recruitment by the μOR, and hold promise for developing next-generation analgesics with fewer adverse effects.
Keywords: Drug Discovery; GPCRs; Medicinal Chemistry; Opioids; cryoEM Structure.
© 2022 The Authors. Angewandte Chemie International Edition published by Wiley-VCH GmbH.
Conflict of interest statement
B.K.K. and P.G. are co‐founders of Epiodyne. B.K.K is a co‐founder of and consultant for ConformetRx.
Figures
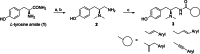


References
-
- Gillis A., Gondin A. B., Kliewer A., Sanchez J., Lim H. D., Alamein C., Manandhar P., Santiago M., Fritzwanker S., Schmidel F., Katte T. A., Reekie T., Grimsey N. L., Kassiou M., Kellam B., Krasel C., Halls M. L., Connor M., Lane J. R., Schulz S., Christie M. J., Canals M., Sci. Signaling 2020, 13, eaaz3140. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

