Augmin-dependent microtubule self-organization drives kinetochore fiber maturation in mammals
- PMID: 35385739
- PMCID: PMC8994134
- DOI: 10.1016/j.celrep.2022.110610
Augmin-dependent microtubule self-organization drives kinetochore fiber maturation in mammals
Abstract
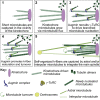
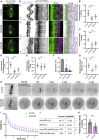
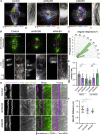
Chromosome segregation in mammals relies on the maturation of a thick bundle of kinetochore-attached microtubules known as k-fiber. How k-fibers mature from initial kinetochore microtubule attachments remains a fundamental question. By combining molecular perturbations and phenotypic analyses in Indian muntjac fibroblasts containing the lowest known diploid chromosome number in mammals (2N = 6) and distinctively large kinetochores, with fixed/live-cell super-resolution coherent-hybrid stimulated emission depletion (CH-STED) nanoscopy and laser microsurgery, we demonstrate a key role for augmin in kinetochore microtubule self-organization and maturation, regardless of pioneer centrosomal microtubules. In doing so, augmin promotes kinetochore and interpolar microtubule turnover and poleward flux. Tracking of microtubule growth events within individual k-fibers reveals a wide angular dispersion, consistent with augmin-mediated branched microtubule nucleation. Augmin depletion reduces the frequency of kinetochore microtubule growth events and hampers efficient repair after acute k-fiber injury by laser microsurgery. Together, these findings underscore the contribution of augmin-mediated microtubule amplification for k-fiber self-organization and maturation in mammals.
Keywords: CP: Cell biology; Indian muntjac; augmin; coherent-hybrid STED; k-fiber; kinetochore; laser microsurgery; microtubules; mitosis; mitotic spindle; super-resolution.
Copyright © 2022 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

