MIMAS: microfluidic platform in tandem with MALDI mass spectrometry for protein quantification from small cell ensembles
- PMID: 35385983
- PMCID: PMC9188328
- DOI: 10.1007/s00216-022-04038-y
MIMAS: microfluidic platform in tandem with MALDI mass spectrometry for protein quantification from small cell ensembles
Abstract
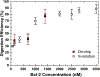
Understanding cell-to-cell variation at the molecular level provides relevant information about biological phenomena and is critical for clinical and biological research. Proteins carry important information not available from single-cell genomics and transcriptomics studies; however, due to the minute amount of proteins in single cells and the complexity of the proteome, quantitative protein analysis at the single-cell level remains challenging. Here, we report an integrated microfluidic platform in tandem with matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF-MS) for the detection and quantification of targeted proteins from small cell ensembles (> 10 cells). All necessary steps for the assay are integrated on-chip including cell lysis, protein immunocapture, tryptic digestion, and co-crystallization with the matrix solution for MALDI-MS analysis. We demonstrate that our approach is suitable for protein quantification by assessing the apoptotic protein Bcl-2 released from MCF-7 breast cancer cells, ranging from 26 to 223 cells lysed on-chip (8.75 nL wells). A limit of detection (LOD) of 11.22 nM was determined, equivalent to 5.91 × 107 protein molecules per well. Additionally, the microfluidic platform design was further improved, establishing the successful quantification of Bcl-2 protein from MCF-7 cell ensembles ranging from 8 to 19 cells in 4 nL wells. The LOD in the smaller well designs for Bcl-2 resulted in 14.85 nM, equivalent to 3.57 × 107 protein molecules per well. This work shows the capability of our approach to quantitatively assess proteins from cell lysate on the MIMAS platform for the first time. These results demonstrate our approach constitutes a promising tool for quantitative targeted protein analysis from small cell ensembles down to single cells, with the capability for multiplexing through parallelization and automation.
Keywords: Mass spectrometry; Microfabrication; Microfluidics; Protein; Quantification.
© 2022. Springer-Verlag GmbH Germany, part of Springer Nature.
Conflict of interest statement
CONFLICT OF INTEREST
The authors declare no competing interests.
Figures

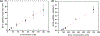



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

