Insights from the rescue and breeding management of Cuvier's gazelle (Gazella cuvieri) through whole-genome sequencing
- PMID: 35386395
- PMCID: PMC8965372
- DOI: 10.1111/eva.13336
Insights from the rescue and breeding management of Cuvier's gazelle (Gazella cuvieri) through whole-genome sequencing
Abstract
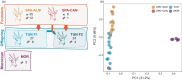
Captive breeding programmes represent the most intensive type of ex situ population management for threatened species. One example is the Cuvier's gazelle programme that started in 1975 with only four founding individuals, and after more than four decades of management in captivity, a reintroduction effort was undertaken in Tunisia in 2016, to establish a population in an area historically included within its range. Here, we aim to determine the genetic consequences of this reintroduction event by assessing the genetic diversity of the founder stock as well as of their descendants. We present the first whole-genome sequencing dataset of 30 Cuvier's gazelles including captive-bred animals, animals born in Tunisia after a reintroduction and individuals from a genetically unrelated Moroccan population. Our analyses revealed no difference between the founder and the offspring cohorts in genome-wide heterozygosity and inbreeding levels, and in the amount and length of runs of homozygosity. The captive but unmanaged Moroccan gazelles have the lowest genetic diversity of all genomes analysed. Our findings demonstrate that the Cuvier's gazelle captive breeding programme can serve as source populations for future reintroductions of this species. We believe that this study can serve as a starting point for global applications of genomics to the conservation plan of this species.
Keywords: Cuvier’s gazelle; captive breeding; genomics; inbreeding; reintroduction.
© 2022 The Authors. Evolutionary Applications published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Andrews, S. (2010). FastQC a quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
-
- Attum, O. , & Mahmoud, T. (2012). Dorcas gazelle and livestock use of trees according to size in a hyper‐arid landscape. Journal of Arid Environments, 76, 49–53. 10.1016/j.jaridenv.2011.07.002 - DOI
-
- Aulagnier, S. , Bayed, A. , Cuzin, F. , & Thévenot, M. (2015). Mammifères du Maroc: extinctions et régressions au cours du XXème siècle. Travaux De L’institut Scientifique, Série Générale, 8, 53–67.
LinkOut - more resources
Full Text Sources

