Estimating the Risk of Severe Peanut Allergy Using Clinical Background and IgE Sensitization Profiles
- PMID: 35386994
- PMCID: PMC8974676
- DOI: 10.3389/falgy.2021.670789
Estimating the Risk of Severe Peanut Allergy Using Clinical Background and IgE Sensitization Profiles
Abstract
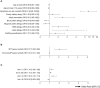
Background: It is not well-understood why symptom severity varies between patients with peanut allergy (PA). Objective: To gain insight into the clinical profile of subjects with mild-to-moderate and severe PA, and investigate individual and collective predictive accuracy of clinical background and IgE to peanut extract and components for PA severity. Methods: Data on demographics, patient history and sensitization at extract and component level of 393 patients with probable PA (symptoms ≤ 2 h + IgE sensitization) from 12 EuroPrevall centers were analyzed. Univariable and penalized multivariable regression analyses were used to evaluate risk factors and biomarkers for severity. Results: Female sex, age at onset of PA, symptoms elicited by skin contact with peanut, family atopy, atopic dermatitis, house dust mite and latex allergy were independently associated with severe PA; birch pollen allergy with mild-to-moderate PA. The cross-validated AUC of all clinical background determinants combined (0.74) was significantly larger than the AUC of tests for sensitization to extract (0.63) or peanut components (0.54-0.64). Although larger skin prick test wheal size, and higher IgE to peanut extract, Ara h 1 and Ara h 2/6, were associated with severe PA, and higher IgE to Ara h 8 with mild-to-moderate PA, addition of these measurements of sensitization to the clinical background model did not significantly improve the AUC. Conclusions: Models combining clinical characteristics and IgE sensitization patterns can help establish the risk of severe reactions for peanut allergic patients, but clinical background determinants are most valuable for predicting severity of probable PA in an individual patient.
Keywords: EuroPrevall; IgE; clinical background; component-resolved diagnostics; iFAAM; peanut allergy; prediction; severity.
Copyright © 2021 Datema, Lyons, Fernández-Rivas, Ballmer-Weber, Knulst, Asero, Barreales, Belohlavkova, de Blay, Clausen, Dubakiene, Fernández-Perez, Fritsche, Gislason, Hoffmann-Sommergruber, Jedrzejczak-Czechowicz, Jongejan, Kowalski, Kralimarkova, Lidholm, Papadopoulos, Popov, Prado, Purohit, Reig, Seneviratne, Sinaniotis, Vassilopoulou, Versteeg, Vieths, Welsing, Mills, Le, Zwinderman and van Ree.
Conflict of interest statement
Outside of submitted work: MF-R reported grants and personal fees from Aimmune Therapeutics and Diater, personal fees from DBV, Allergy Therapeutics, GSK, HAL Allergy, Novartis, ThermoFisher Scientific, and SPRIM. BB-W reported personal fees from ThermoFisher Scientific. FB reported personal fees from Aimmune; grants from Stallergènes Greer, Chiesi, Mundipharma, Novartis, and Regeneron; and board membership for DVB, Stallergènes Greer, Novartis, ALK, Mundipharma, Boehringer, AstraZeneca, Medapharma, and Boston Scientific. JL was an employee of ThermoFisher Scientific. NP reported personal fees from Novartis, Nutricia, HAL Allergy BV, Menarine/Faes Farma, Sanofi, Mylan/Meda, Biomay, AstraZeneca, GSK, MSD, ASIT Biotech, Boehringer Ingelheim; and grants from Gerolymatos International SA, and Capricare. SV reported personal fees from Ärzteverband Deutscher Allergologen, Swiss Society for Allergy and Immunology, Schattauer Allergologie Handbuch, Elsevier Nahrungsmittelallergien und Intoleranzen, Karger Food Allergy: Molecular Basis and Clinical Practice, and Pharmacon. EM reported grants from Reacta Biotech; and was shareholder of Reacta Biotech Ltd. RR reported personal fees from HAL Allergy BV, Citeq BV, Angany Inc., and ThermoFisher Scientific. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
LinkOut - more resources
Full Text Sources

