High quality genome assembly of the anhydrobiotic midge provides insights on a single chromosome-based emergence of extreme desiccation tolerance
- PMID: 35387384
- PMCID: PMC8982440
- DOI: 10.1093/nargab/lqac029
High quality genome assembly of the anhydrobiotic midge provides insights on a single chromosome-based emergence of extreme desiccation tolerance
Abstract
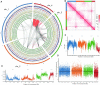
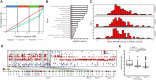
Non-biting midges (Chironomidae) are known to inhabit a wide range of environments, and certain species can tolerate extreme conditions, where the rest of insects cannot survive. In particular, the sleeping chironomid Polypedilum vanderplanki is known for the remarkable ability of its larvae to withstand almost complete desiccation by entering a state called anhydrobiosis. Chromosome numbers in chironomids are higher than in other dipterans and this extra genomic resource might facilitate rapid adaptation to novel environments. We used improved sequencing strategies to assemble a chromosome-level genome sequence for P. vanderplanki for deep comparative analysis of genomic location of genes associated with desiccation tolerance. Using whole genome-based cross-species and intra-species analysis, we provide evidence for the unique functional specialization of Chromosome 4 through extensive acquisition of novel genes. In contrast to other insect genomes, in the sleeping chironomid a uniquely high degree of subfunctionalization in paralogous anhydrobiosis genes occurs in this chromosome, as well as pseudogenization in a highly duplicated gene family. Our findings suggest that the Chromosome 4 in Polypedilum is a site of high genetic turnover, allowing it to act as a 'sandbox' for evolutionary experiments, thus facilitating the rapid adaptation of midges to harsh environments.
© The Author(s) 2022. Published by Oxford University Press on behalf of NAR Genomics and Bioinformatics.
Figures

References
-
- Pinder L.C.V. Biology of freshwater chironomidae. Annu. Rev. Entomol. 1986; 31:1–23.
-
- Armitage P.D., Cranston P.S., Pinder L.C.V. The Chironomidae. 1995; Springer, Dordrecht, Netherlands.
-
- Hinton H.E. Cryptobiosis in the larva of Polypedilum vanderplanki Hint. (Chironomidae). J. Insect Physiol. 1960; 5:286–300.
-
- Watanabe M., Kikawada T., Minagawa N., Yukuhiro F., Okuda T. Mechanism allowing an insect to survive complete dehydration and extreme temperatures. J. Exp. Biol. 2002; 205:2799–2802. - PubMed
-
- Watanabe M., Kikawada T., Okuda T. Increase of internal ion concentration triggers trehalose synthesis associated with cryptobiosis in larvae of Polypedilum vanderplanki. J. Exp. Biol. 2003; 206:2281–2286. - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

