Implementation of Zebrafish Ontologies for Toxicology Screening
- PMID: 35387429
- PMCID: PMC8979167
- DOI: 10.3389/ftox.2022.817999
Implementation of Zebrafish Ontologies for Toxicology Screening
Abstract
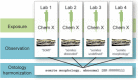
Toxicological evaluation of chemicals using early-life stage zebrafish (Danio rerio) involves the observation and recording of altered phenotypes. Substantial variability has been observed among researchers in phenotypes reported from similar studies, as well as a lack of consistent data annotation, indicating a need for both terminological and data harmonization. When examined from a data science perspective, many of these apparent differences can be parsed into the same or similar endpoints whose measurements differ only in time, methodology, or nomenclature. Ontological knowledge structures can be leveraged to integrate diverse data sets across terminologies, scales, and modalities. Building on this premise, the National Toxicology Program's Systematic Evaluation of the Application of Zebrafish in Toxicology undertook a collaborative exercise to evaluate how the application of standardized phenotype terminology improved data consistency. To accomplish this, zebrafish researchers were asked to assess images of zebrafish larvae for morphological malformations in two surveys. In the first survey, researchers were asked to annotate observed malformations using their own terminology. In the second survey, researchers were asked to annotate the images from a list of terms and definitions from the Zebrafish Phenotype Ontology. Analysis of the results suggested that the use of ontology terms increased consistency and decreased ambiguity, but a larger study is needed to confirm. We conclude that utilizing a common data standard will not only reduce the heterogeneity of reported terms but increases agreement and repeatability between different laboratories. Thus, we advocate for the development of a zebrafish phenotype atlas to help laboratories create interoperable, computable data.
Keywords: Danio rerio; annotation; endpoint; ontology; phenotype; zebrafish.
Copyright © 2022 Thessen, Marvel, Achenbach, Fischer, Haendel, Hayward, Klüver, Könemann, Legradi, Lein, Leong, Mylroie, Padilla, Perone, Planchart, Prieto, Muriana, Quevedo, Reif, Ryan, Stinckens, Truong, Vergauwen, Vom Berg, Wilbanks, Yaghoobi and Hamm.
Conflict of interest statement
Author SF is employed by aQuaTox-Solutions Ltd. Author AM is employed by Biobide. Author CQ is employed by Viralgen Vector Core. Author JH is employed by Integrated Laboratory Systems, LLC. Author RP is employed by ZeClinics SL. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures







References
-
- Boyles R. R., Thessen A. E., Waldrop A., Haendel M. A. (2019). Ontology-based Data Integration for Advancing Toxicological Knowledge. Curr. Opin. Toxicol. 16, 67–74. 10.1016/j.cotox.2019.05.005 - DOI
LinkOut - more resources
Full Text Sources

