Common and Rare 5'UTR Variants Altering Upstream Open Reading Frames in Cardiovascular Genomics
- PMID: 35387445
- PMCID: PMC8977850
- DOI: 10.3389/fcvm.2022.841032
Common and Rare 5'UTR Variants Altering Upstream Open Reading Frames in Cardiovascular Genomics
Abstract
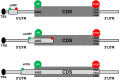
High-throughput sequencing (HTS) technologies are revolutionizing the research and molecular diagnosis landscape by allowing the exploration of millions of nucleotide sequences at an unprecedented scale. These technologies are of particular interest in the identification of genetic variations contributing to the risk of rare (Mendelian) and common (multifactorial) human diseases. So far, they have led to numerous successes in identifying rare disease-causing mutations in coding regions, but few in non-coding regions that include introns, untranslated (UTR), and intergenic regions. One class of neglected non-coding variations is that of 5'UTR variants that alter upstream open reading frames (upORFs) of the coding sequence (CDS) of a natural protein coding transcript. Following a brief summary of the molecular bases of the origin and functions of upORFs, we will first review known 5'UTR variations altering upORFs and causing rare cardiovascular disorders (CVDs). We will then investigate whether upORF-affecting single nucleotide polymorphisms could be good candidates for explaining association signals detected in the context of genome-wide association studies for common complex CVDs.
Keywords: Mendelian disease; genome wide association analysis (GWAS); non-coding mutations; open reading frame (ORF); polymorphism.
Copyright © 2022 Soukarieh, Meguerditchian, Proust, Aïssi, Eyries, Goyenvalle and Trégouët.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

