Linking transcriptional dynamics of CH4-cycling grassland soil microbiomes to seasonal gas fluxes
- PMID: 35388141
- PMCID: PMC9213473
- DOI: 10.1038/s41396-022-01229-4
Linking transcriptional dynamics of CH4-cycling grassland soil microbiomes to seasonal gas fluxes
Abstract
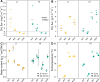
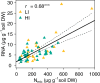
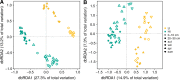
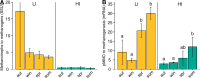
Soil CH4 fluxes are driven by CH4-producing and -consuming microorganisms that determine whether soils are sources or sinks of this potent greenhouse gas. To date, a comprehensive understanding of underlying microbiome dynamics has rarely been obtained in situ. Using quantitative metatranscriptomics, we aimed to link CH4-cycling microbiomes to net surface CH4 fluxes throughout a year in two grassland soils. CH4 fluxes were highly dynamic: both soils were net CH4 sources in autumn and winter and sinks in spring and summer, respectively. Correspondingly, methanogen mRNA abundances per gram soil correlated well with CH4 fluxes. Methanotroph to methanogen mRNA ratios were higher in spring and summer, when the soils acted as net CH4 sinks. CH4 uptake was associated with an increased proportion of USCα and γ pmoA and pmoA2 transcripts. We assume that methanogen transcript abundance may be useful to approximate changes in net surface CH4 emissions from grassland soils. High methanotroph to methanogen ratios would indicate CH4 sink properties. Our study links for the first time the seasonal transcriptional dynamics of CH4-cycling soil microbiomes to gas fluxes in situ. It suggests mRNA transcript abundances as promising indicators of dynamic ecosystem-level processes.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Canadell JG, Monteiro PMS, Costa, MH, Cotrim da Cunha L, Cox PM, Eliseev AV, et al. Global carbon and other biogeochemical cycles and feedbacks. In: Masson-Delmotte V, Zhai P, Pirani A, Connors SL, Péan C, Berger S, et al. editors. Climate Change 2021: The Physical Science Basis. Contribution of Working Group I to the Sixth Assessment Report of the Intergovernmental Panel on Climate Change. Cambridge University Press; 2021, in press.
-
- Rosentreter JA, Borges AV, Deemer BR, Holgerson MA, Liu S, Song C, et al. Half of global methane emissions come from highly variable aquatic ecosystem sources. Nat Geosci. 2021;14:225–30. doi: 10.1038/s41561-021-00715-2. - DOI
-
- Saunois M, Stavert AR, Poulter B, Bousquet P, Canadell JG, Jackson RB, et al. The global methane budget 2000 – 2017. Earth Syst. Sci Data. 2020;12:1561–623.
-
- Lamentowicz M, Gałka M, Pawlyta J, Lamentowicz Ł, Goslar T, Miotk-Szpiganowicz G. Climate change and human impact in the southern Baltic during the last millennium reconstructed from an ombrotrophic bog archive. Stud Quat. 2011;28:3–16.
-
- Davidson NC. How much wetland has the world lost? Long-term and recent trends in global wetland area. Mar Freshw Res. 2014;65:934–41. doi: 10.1071/MF14173. - DOI

