Two defence systems eliminate plasmids from seventh pandemic Vibrio cholerae
- PMID: 35388218
- PMCID: PMC7613841
- DOI: 10.1038/s41586-022-04546-y
Two defence systems eliminate plasmids from seventh pandemic Vibrio cholerae
Abstract
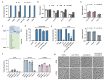
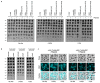
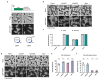
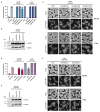
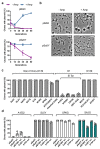
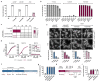
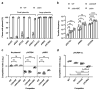
Horizontal gene transfer can trigger rapid shifts in bacterial evolution. Driven by a variety of mobile genetic elements-in particular bacteriophages and plasmids-the ability to share genes within and across species underpins the exceptional adaptability of bacteria. Nevertheless, invasive mobile genetic elements can also present grave risks to the host; bacteria have therefore evolved a vast array of defences against these elements1. Here we identify two plasmid defence systems conserved in the Vibrio cholerae El Tor strains responsible for the ongoing seventh cholera pandemic2-4. These systems, termed DdmABC and DdmDE, are encoded on two major pathogenicity islands that are a hallmark of current pandemic strains. We show that the modules cooperate to rapidly eliminate small multicopy plasmids by degradation. Moreover, the DdmABC system is widespread and can defend against bacteriophage infection by triggering cell suicide (abortive infection, or Abi). Notably, we go on to show that, through an Abi-like mechanism, DdmABC increases the burden of large low-copy-number conjugative plasmids, including a broad-host IncC multidrug resistance plasmid, which creates a fitness disadvantage that counterselects against plasmid-carrying cells. Our results answer the long-standing question of why plasmids, although abundant in environmental strains, are rare in pandemic strains; have implications for understanding the dissemination of antibiotic resistance plasmids; and provide insights into how the interplay between two defence systems has shaped the evolution of the most successful lineage of pandemic V. cholerae.
© 2022. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
M.B, M.J, and D.W.A. are inventors on the patent application EP21183501.2 filed with the European Patent Office.
Figures







Comment in
-
Cholera-causing bacteria have defences that degrade plasmid invaders.Nature. 2022 Apr;604(7905):250-252. doi: 10.1038/d41586-022-00871-4. Nature. 2022. PMID: 35388150 No abstract available.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

