Compartmentalized metabolism supports midgestation mammalian development
- PMID: 35388219
- PMCID: PMC9007737
- DOI: 10.1038/s41586-022-04557-9
Compartmentalized metabolism supports midgestation mammalian development
Abstract
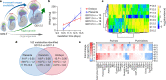
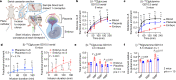
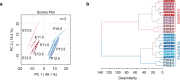
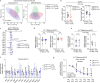
Mammalian embryogenesis requires rapid growth and proper metabolic regulation1. Midgestation features increasing oxygen and nutrient availability concomitant with fetal organ development2,3. Understanding how metabolism supports development requires approaches to observe metabolism directly in model organisms in utero. Here we used isotope tracing and metabolomics to identify evolving metabolic programmes in the placenta and embryo during midgestation in mice. These tissues differ metabolically throughout midgestation, but we pinpointed gestational days (GD) 10.5-11.5 as a transition period for both placenta and embryo. Isotope tracing revealed differences in carbohydrate metabolism between the tissues and rapid glucose-dependent purine synthesis, especially in the embryo. Glucose's contribution to the tricarboxylic acid (TCA) cycle rises throughout midgestation in the embryo but not in the placenta. By GD12.5, compartmentalized metabolic programmes are apparent within the embryo, including different nutrient contributions to the TCA cycle in different organs. To contextualize developmental anomalies associated with Mendelian metabolic defects, we analysed mice deficient in LIPT1, the enzyme that activates 2-ketoacid dehydrogenases related to the TCA cycle4,5. LIPT1 deficiency suppresses TCA cycle metabolism during the GD10.5-GD11.5 transition, perturbs brain, heart and erythrocyte development and leads to embryonic demise by GD11.5. These data document individualized metabolic programmes in developing organs in utero.
© 2022. The Author(s).
Conflict of interest statement
R.J.D. is an advisor for Agios Pharmaceuticals and Vida Ventures and a co-founder of Atavistik Bio. The other authors declare no competing interests.
Figures







References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

