An overview of gene regulation in bacteria by small RNAs derived from mRNA 3' ends
- PMID: 35388892
- PMCID: PMC9438474
- DOI: 10.1093/femsre/fuac017
An overview of gene regulation in bacteria by small RNAs derived from mRNA 3' ends
Abstract
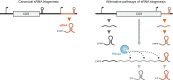
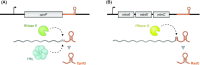
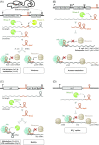
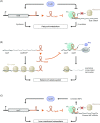
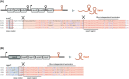
Over the past two decades, small noncoding RNAs (sRNAs) that regulate mRNAs by short base pairing have gone from a curiosity to a major class of post-transcriptional regulators in bacteria. They are integral to many stress responses and regulatory circuits, affecting almost all aspects of bacterial life. Following pioneering sRNA searches in the early 2000s, the field quickly focused on conserved sRNA genes in the intergenic regions of bacterial chromosomes. Yet, it soon emerged that there might be another rich source of bacterial sRNAs-processed 3' end fragments of mRNAs. Several such 3' end-derived sRNAs have now been characterized, often revealing unexpected, conserved functions in diverse cellular processes. Here, we review our current knowledge of these 3' end-derived sRNAs-their biogenesis through ribonucleases, their molecular mechanisms, their interactions with RNA-binding proteins such as Hfq or ProQ and their functional scope, which ranges from acting as specialized regulators of single metabolic genes to constituting entire noncoding arms in global stress responses. Recent global RNA interactome studies suggest that the importance of functional 3' end-derived sRNAs has been vastly underestimated and that this type of cross-regulation between genes at the mRNA level is more pervasive in bacteria than currently appreciated.
Keywords: 3′ UTR; bacteria; post-transcriptional control; regulatory networks; sRNA.
© The Author(s) 2022. Published by Oxford University Press on behalf of FEMS.
Figures




References
-
- Acuña LG, Barros MJ, Peñaloza Det al. A feed-forward loop between SroC and MgrR small RNAs modulates the expression of eptB and the susceptibility to polymyxin B in Salmonella Typhimurium. Microbiology. 2016;162:1996–2004. - PubMed
-
- Ahn BE, Cha J, Lee EJet al. Nur, a nickel-responsive regulator of the Fur family, regulates superoxide dismutases and nickel transport in Streptomyces coelicolor. Mol Microbiol. 2006;59:1848–58. - PubMed
-
- Argaman L, Hershberg R, Vogel Jet al. Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr Biol. 2001;11:941–50. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

