MOSPD2 is an endoplasmic reticulum-lipid droplet tether functioning in LD homeostasis
- PMID: 35389430
- PMCID: PMC8996327
- DOI: 10.1083/jcb.202110044
MOSPD2 is an endoplasmic reticulum-lipid droplet tether functioning in LD homeostasis
Abstract
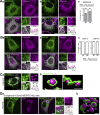
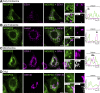
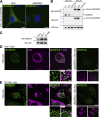
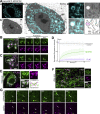
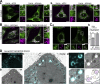
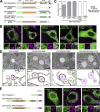
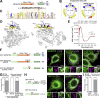
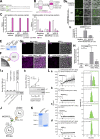
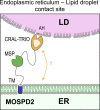
Membrane contact sites between organelles are organized by protein bridges. Among the components of these contacts, the VAP family comprises ER-anchored proteins, such as MOSPD2, that function as major ER-organelle tethers. MOSPD2 distinguishes itself from the other members of the VAP family by the presence of a CRAL-TRIO domain. In this study, we show that MOSPD2 forms ER-lipid droplet (LD) contacts, thanks to its CRAL-TRIO domain. MOSPD2 ensures the attachment of the ER to LDs through a direct protein-membrane interaction. The attachment mechanism involves an amphipathic helix that has an affinity for lipid packing defects present at the surface of LDs. Remarkably, the absence of MOSPD2 markedly disturbs the assembly of lipid droplets. These data show that MOSPD2, in addition to being a general ER receptor for inter-organelle contacts, possesses an additional tethering activity and is specifically implicated in the biology of LDs via its CRAL-TRIO domain.
© 2022 ZOUIOUICH et al.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

