Polyploidy on islands - concerted evolution and gene loss amid chromosomal stasis
- PMID: 35390127
- PMCID: PMC9904340
- DOI: 10.1093/aob/mcac051
Polyploidy on islands - concerted evolution and gene loss amid chromosomal stasis
Abstract
Background and aims: Polyploidy is an important process that often generates genomic diversity within lineages, but it can also cause changes that result in loss of genomic material. Island lineages, while often polyploid, typically show chromosomal stasis but have not been investigated in detail regarding smaller-scale gene loss. Our aim was to investigate post-polyploidization genome dynamics in a chromosomally stable lineage of Malvaceae endemic to New Zealand.
Methods: We determined chromosome numbers and used fluorescence in situ hybridization to localize 18S and 5S rDNA. Gene sequencing of 18S rDNA, the internal transcribed spacers (ITS) with intervening 5.8S rDNA, and a low-copy nuclear gene, GBSSI-1, was undertaken to determine if gene loss occurred in the New Zealand lineage following polyploidy.
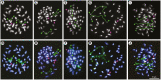
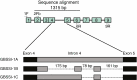
Key results: The chromosome number for all species investigated was 2n = 42, with the first published report for the monotypic Australian genus Asterotrichion. The five species investigated all had two 5S rDNA signals localized interstitially on the long arm of one of the largest chromosome pairs. All species, except Plagianthus regius, had two 18S rDNA signals localized proximally on the short arm of one of the smallest chromosome pairs. Plagianthus regius had two additional 18S rDNA signals on a separate chromosome, giving a total of four. Sequencing of nuclear ribosomal 18S rDNA and the ITS cistron indicated loss of historical ribosomal repeats. Phylogenetic analysis of a low-copy nuclear gene, GBSSI-1, indicated that some lineages maintained three copies of the locus, while others have lost one or two copies.
Conclusions: Although island endemic lineages show chromosomal stasis, with no additional changes in chromosome number, they may undergo smaller-scale processes of gene loss and concerted evolution ultimately leading to further genome restructuring and downsizing.
Keywords: Plagianthus alliance; 18S rDNA; GBSSI; ITS; Malvaceae; chromosome number; concerted evolution; fluorescence in situ hybridization; gene fractionation; polyploid; rDNA.
© The Author(s) 2022. Published by Oxford University Press on behalf of the Annals of Botany Company.
Figures




References
-
- Alvarez I, Wendel JF. 2003. Ribosomal ITS sequences and plant phylogenetic inference. Molecular Phylogenetics & Evolution 29: 417–434. - PubMed
-
- Ansari HA, Ellison NW, Reader SM, et al. 1999. Molecular cytogenetic organization of 5S and 18S-26S rDNA loci in white clover (Trifolium repens L.) and related species. Annals of Botany 83: 199–206.
-
- Ansari H, Ellison N, Williams W. 2008. Molecular and cytogenetic evidence for an allotetraploid origin of Trifolium dubium (Leguminosae). Chromosoma 117: 159–167. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

