CB2R Activation Regulates TFEB-Mediated Autophagy and Affects Lipid Metabolism and Inflammation of Astrocytes in POCD
- PMID: 35392078
- PMCID: PMC8981088
- DOI: 10.3389/fimmu.2022.836494
CB2R Activation Regulates TFEB-Mediated Autophagy and Affects Lipid Metabolism and Inflammation of Astrocytes in POCD
Abstract
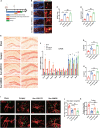
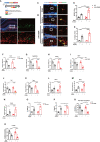
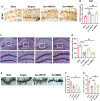
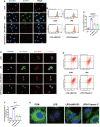
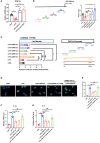
Evidence suggests that the accumulation of lipid drots (LDs) accelerates damage to mitochondria and increases the release of inflammatory factors. These have been implicated as a mechanism underlying neurodegenerative diseases or tumors and aging-related diseases such as postoperative cognitive dysfunction (POCD), nevertheless, accumulation of lipid droplets has not been extensively studied in the central nervous system (CNS). Here, we found that after surgery, there was activation of astrocytes and lipid accumulation in the hippocampus. However, cannabinoid receptor type II (CB2R) activation significantly reduced lipid accumulation in astrocytes and change the expression of genes related to lipid metabolism. CB2R reduces the release of the inflammatory factors interleukin-1 beta (IL-1β) and interleukin 6 (IL-6) in peripheral serum and simultaneously improves cognitive ability in mice with POCD. Further research on mechanisms indicates that CB2R activation promotes the nuclear entry of the bHLH-leucine zipper transcription factor, the transcription factor EB (TFEB), and which is a master transcription factor of the autophagy-lysosomal pathway, also reduces TFEB-S211 phosphorylation. When CB2R promotes TFEB into the nucleus, TFEB binds at two sites within promoter region of PGC1α, promoting PGC1α transcription and accelerating downstream lipid metabolism. The aforementioned process leads to autophagy activation and decreases cellular lipid content. This study uncovers a new mechanism allowing CB2R to regulate lipid metabolism and inflammation in POCD.
Keywords: astrocytes; autophagy; cannabinoid type 2 receptor; inflammation; lipid accumulation; mitochondrial damage; postoperative cognitive dysfunction.
Copyright © 2022 Zhang, Wang, Yu, Ying, Fang, Zheng, Feng, Hu, Xiao, Chen, Wei, Lin, Liu, Yang, Fang, Xu and Hua.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Activation of peroxisome proliferator-activated receptor α induces lysosomal biogenesis in brain cells: implications for lysosomal storage disorders.J Biol Chem. 2015 Apr 17;290(16):10309-24. doi: 10.1074/jbc.M114.610659. Epub 2015 Mar 6. J Biol Chem. 2015. PMID: 25750174 Free PMC article.
-
Impaired TFEB-mediated lysosomal biogenesis promotes the development of pancreatitis in mice and is associated with human pancreatitis.Autophagy. 2019 Nov;15(11):1954-1969. doi: 10.1080/15548627.2019.1596486. Epub 2019 Mar 30. Autophagy. 2019. PMID: 30894069 Free PMC article.
-
Mitochondrial Quality Control via the PGC1α-TFEB Signaling Pathway Is Compromised by Parkin Q311X Mutation But Independently Restored by Rapamycin.J Neurosci. 2015 Sep 16;35(37):12833-44. doi: 10.1523/JNEUROSCI.0109-15.2015. J Neurosci. 2015. PMID: 26377470 Free PMC article.
-
TFEB at a glance.J Cell Sci. 2016 Jul 1;129(13):2475-81. doi: 10.1242/jcs.146365. Epub 2016 Jun 1. J Cell Sci. 2016. PMID: 27252382 Free PMC article. Review.
-
TFEB dysregulation as a driver of autophagy dysfunction in neurodegenerative disease: Molecular mechanisms, cellular processes, and emerging therapeutic opportunities.Neurobiol Dis. 2019 Feb;122:83-93. doi: 10.1016/j.nbd.2018.05.012. Epub 2018 May 28. Neurobiol Dis. 2019. PMID: 29852219 Free PMC article. Review.
Cited by
-
A Proteomic Approach Identified TFEB as a Key Player in the Protective Action of Novel CB2R Bitopic Ligand FD22a against the Deleterious Effects Induced by β-Amyloid in Glial Cells.Cells. 2024 May 19;13(10):875. doi: 10.3390/cells13100875. Cells. 2024. PMID: 38786097 Free PMC article.
-
Astrocyte mitochondria: Potential therapeutic targets for epilepsy.Heliyon. 2024 Apr 27;10(9):e29950. doi: 10.1016/j.heliyon.2024.e29950. eCollection 2024 May 15. Heliyon. 2024. PMID: 38756598 Free PMC article. Review.
-
The Role of Astrocytes in the Mechanism of Perioperative Neurocognitive Disorders.Brain Sci. 2022 Oct 25;12(11):1435. doi: 10.3390/brainsci12111435. Brain Sci. 2022. PMID: 36358362 Free PMC article.
-
Genome-wide DNA methylation association study of recent and cumulative marijuana use in middle aged adults.Mol Psychiatry. 2023 Jun;28(6):2572-2582. doi: 10.1038/s41380-023-02106-y. Epub 2023 May 31. Mol Psychiatry. 2023. PMID: 37258616 Free PMC article.
-
Transcutaneous electrical acupoint stimulation for reducing cognitive dysfunction in lumbar spine surgery: A randomized, controlled trail.Front Aging Neurosci. 2022 Dec 5;14:1034998. doi: 10.3389/fnagi.2022.1034998. eCollection 2022. Front Aging Neurosci. 2022. PMID: 36545028 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

