An Easy and Rapid Transformation Protocol for Transient Expression in Cotton Fiber
- PMID: 35392510
- PMCID: PMC8980934
- DOI: 10.3389/fpls.2022.837994
An Easy and Rapid Transformation Protocol for Transient Expression in Cotton Fiber
Abstract
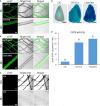
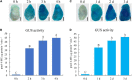
Cotton fiber is the most important natural textile material in the world. Identification and functional characterization of genes regulating fiber development are fundamental for improving fiber quality and yield. However, stable cotton transformation is time-consuming, low in efficiency, and technically complex. Moreover, heterologous systems, such as Arabidopsis and tobacco, did not always work to elucidate the function of cotton fiber specifically expressed genes or their promoters. For these reasons, constructing a rapid transformation system using cotton fibers is necessary to study fiber's specifically expressed genes. In this study, we developed an easy and rapid Agrobacterium-mediated method for the transient transformation of genes and promoters in cotton fibers. First, we found that exogenous genes could be expressed in cotton fibers via using β-glucuronidase (GUS) and green fluorescence protein (GFP) as reporters. Second, parameters affecting transformation efficiency, including LBA4404 Agrobacterium strain, 3 h infection time, and 2-day incubation time, were determined. Third, four different cotton genes that are specifically expressed in fibers were transiently transformed in cotton fibers, and the transcripts of these genes were detected ten to thousand times increase over the control. Fourth, GUS staining and activity analysis demonstrated that the activity profiles of GhMYB212 and GhFSN1 promoters in transformed fibers are similar to their native activity in developmental fibers. Furthermore, the transient transformation method was confirmed to be suitable for subcellular localization studies. In summary, the presented Agrobacterium-mediated transient transformation method is a fast, simple, and effective system for promoter characterization and protein expression in cotton fibers.
Keywords: Agrobacterium; cotton fiber; promoter activity; subcellular localization; transient transformation.
Copyright © 2022 Shang, Zhu, Duan, He, Zhao, Yu and Guo.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
A transient transformation system for gene characterization in upland cotton (Gossypium hirsutum).Plant Methods. 2018 Jun 22;14:50. doi: 10.1186/s13007-018-0319-2. eCollection 2018. Plant Methods. 2018. PMID: 29977323 Free PMC article.
-
The cotton (Gossypium hirsutum) NAC transcription factor (FSN1) as a positive regulator participates in controlling secondary cell wall biosynthesis and modification of fibers.New Phytol. 2018 Jan;217(2):625-640. doi: 10.1111/nph.14864. Epub 2017 Nov 6. New Phytol. 2018. PMID: 29105766
-
An improved and efficient method of Agrobacterium syringe infiltration for transient transformation and its application in the elucidation of gene function in poplar.BMC Plant Biol. 2021 Jan 21;21(1):54. doi: 10.1186/s12870-021-02833-w. BMC Plant Biol. 2021. PMID: 33478390 Free PMC article.
-
A high-efficiency Agrobacterium-mediated transient expression system in the leaves of Artemisia annua L.Plant Methods. 2021 Oct 16;17(1):106. doi: 10.1186/s13007-021-00807-5. Plant Methods. 2021. PMID: 34654448 Free PMC article.
-
Improvement of cotton fiber quality by transforming the acsA and acsB genes into Gossypium hirsutum L. by means of vacuum infiltration.Plant Cell Rep. 2004 Apr;22(9):691-7. doi: 10.1007/s00299-003-0751-1. Epub 2004 Jan 23. Plant Cell Rep. 2004. PMID: 14740167
Cited by
-
Revealing Genetic Differences in Fiber Elongation between the Offspring of Sea Island Cotton and Upland Cotton Backcross Populations Based on Transcriptome and Weighted Gene Coexpression Networks.Genes (Basel). 2022 May 26;13(6):954. doi: 10.3390/genes13060954. Genes (Basel). 2022. PMID: 35741716 Free PMC article.
-
Establishment and Validation of an Efficient Agrobacterium Tumefaciens-Mediated Transient Transformation System for Salix Psammophila.Int J Mol Sci. 2024 Dec 1;25(23):12934. doi: 10.3390/ijms252312934. Int J Mol Sci. 2024. PMID: 39684643 Free PMC article.
-
The establishment of transient expression systems and their application for gene function analysis of flavonoid biosynthesis in Carthamus tinctorius L.BMC Plant Biol. 2023 Apr 10;23(1):186. doi: 10.1186/s12870-023-04210-1. BMC Plant Biol. 2023. PMID: 37032332 Free PMC article.
References
-
- Chen H., Nelson R. S., Sherwood J. L. (1994). Enhanced recovery of transformants of Agrobacterium tumefaciens after freeze-thaw transformation and drug selection. Biotechniques 16:670. - PubMed
-
- D’Aoust M. A., Lavoie P. O., Couture M. M. J., Trépanier S., Guay J.-M., Dargis M., et al. (2008). Influenza virus-like particles produced by transient expression in nicotiana benthamiana induce a protective immune response against a lethal viral challenge in mice. Plant Biotechnol. J. 6 930–940. 10.1111/j.1467-7652.2008.00384 - DOI - PubMed
LinkOut - more resources
Full Text Sources

