Genome-Wide Analysis of Late Embryogenesis Abundant Protein Gene Family in Vigna Species and Expression of VrLEA Encoding Genes in Vigna glabrescens Reveal Its Role in Heat Tolerance
- PMID: 35392521
- PMCID: PMC8981728
- DOI: 10.3389/fpls.2022.843107
Genome-Wide Analysis of Late Embryogenesis Abundant Protein Gene Family in Vigna Species and Expression of VrLEA Encoding Genes in Vigna glabrescens Reveal Its Role in Heat Tolerance
Abstract
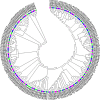
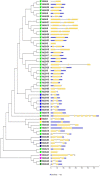
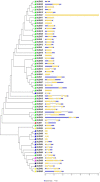
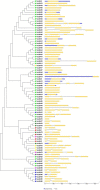
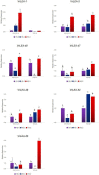
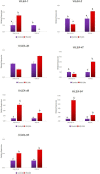
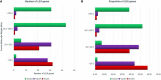
Late embryogenesis abundant (LEA) proteins are identified in many crops for their response and role in adaptation to various abiotic stresses, such as drought, salinity, and temperature. The LEA genes have been studied systematically in several crops but not in Vigna crops. In this study, we reported the first comprehensive analysis of the LEA gene family in three legume species, namely, mung bean (Vigna radiata), adzuki bean (Vigna angularis), and cowpea (Vigna unguiculata), and the cross-species expression of VrLEA genes in a wild tetraploid species, Vigna glabrescens. A total of 201 LEA genes from three Vigna crops were identified harboring the LEA conserved motif. Among these 55, 64, and 82 LEA genes were identified in mung bean, adzuki bean, and cowpea genomes, respectively. These LEA genes were grouped into eight different classes. Our analysis revealed that the cowpea genome comprised all eight classes of LEA genes, whereas the LEA-6 class was absent in the mung bean genome. Similarly, LEA-5 and LEA-6 were absent in the adzuki bean genome. The analysis of LEA genes provides an insight into their structural and functional diversity in the Vigna genome. The genes, such as VrLEA-2, VrLEA-40, VrLEA-47, and VrLEA-55, were significantly upregulated in the heat-tolerant genotype under stress conditions indicating the basis of heat tolerance. The successful amplification and expression of VrLEA genes in V. glabrescens indicated the utility of the developed markers in mung bean improvement. The results of this study increase our understanding of LEA genes and provide robust candidate genes for future functional investigations and a basis for improving heat stress tolerance in Vigna crops.
Keywords: LEA genes; abiotic stress; candidate genes; expression analysis; heat stress; mung bean; wild Vigna.
Copyright © 2022 Singh, Kumar, Pratap, Tripathi, Singh, Mishra, Kumar, Nair and Singh.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Genome-wide analysis of OSCA gene family members in Vigna radiata and their involvement in the osmotic response.BMC Plant Biol. 2021 Sep 7;21(1):408. doi: 10.1186/s12870-021-03184-2. BMC Plant Biol. 2021. PMID: 34493199 Free PMC article.
-
Integrating genetic maps in bambara groundnut [Vigna subterranea (L) Verdc.] and their syntenic relationships among closely related legumes.BMC Genomics. 2017 Feb 20;18(1):192. doi: 10.1186/s12864-016-3393-8. BMC Genomics. 2017. PMID: 28219341 Free PMC article.
-
Genome-wide identification, molecular evolution and expression analysis of the B-box gene family in mung bean (Vigna radiata L.).BMC Plant Biol. 2024 Jun 12;24(1):532. doi: 10.1186/s12870-024-05236-9. BMC Plant Biol. 2024. PMID: 38862892 Free PMC article.
-
Adzuki bean (Vigna angularis): Chemical compositions, physicochemical properties, health benefits, and food applications.Compr Rev Food Sci Food Saf. 2022 May;21(3):2335-2362. doi: 10.1111/1541-4337.12945. Epub 2022 Apr 1. Compr Rev Food Sci Food Saf. 2022. PMID: 35365946 Review.
-
The Potential of the Adzuki Bean (Vigna angularis) and Its Bioactive Compounds in Managing Type 2 Diabetes and Glucose Metabolism: A Narrative Review.Nutrients. 2024 Jan 22;16(2):329. doi: 10.3390/nu16020329. Nutrients. 2024. PMID: 38276567 Free PMC article. Review.
Cited by
-
Insights into morphological and physio-biochemical adaptive responses in mungbean (Vigna radiata L.) under heat stress.Front Genet. 2023 Jun 15;14:1206451. doi: 10.3389/fgene.2023.1206451. eCollection 2023. Front Genet. 2023. PMID: 37396038 Free PMC article. Review.
-
Identification of heat-tolerant mungbean genotypes through morpho-physiological evaluation and key gene expression analysis.Front Genet. 2024 Oct 10;15:1482956. doi: 10.3389/fgene.2024.1482956. eCollection 2024. Front Genet. 2024. PMID: 39449825 Free PMC article.
-
Genome-wide identification, comprehensive characterization of transcription factors, cis-regulatory elements, protein homology, and protein interaction network of DREB gene family in Solanum lycopersicum.Front Plant Sci. 2022 Nov 24;13:1031679. doi: 10.3389/fpls.2022.1031679. eCollection 2022. Front Plant Sci. 2022. PMID: 36507398 Free PMC article.
-
Genome-wide identification and characterization of Subtilisin-like Serine protease encoding genes in Vigna radiata L. Wilczek.Sci Rep. 2025 Apr 17;15(1):13284. doi: 10.1038/s41598-025-95331-0. Sci Rep. 2025. PMID: 40246940 Free PMC article.
-
Exploiting genetic and genomic resources to enhance productivity and abiotic stress adaptation of underutilized pulses.Front Genet. 2023 Jun 16;14:1193780. doi: 10.3389/fgene.2023.1193780. eCollection 2023. Front Genet. 2023. PMID: 37396035 Free PMC article. Review.
References
-
- Allito B. B., Nana E. M., Alemneh A. A. (2015). Rhizobia strain and legume genome interaction effects on nitrogen fixation and yield of grain legume: a review. Mol. Soil Biol. 2 1–6. 10.5376/msb.2015.06.0002 - DOI
LinkOut - more resources
Full Text Sources

