EFG1, Everyone's Favorite Gene in Candida albicans: A Comprehensive Literature Review
- PMID: 35392604
- PMCID: PMC8980467
- DOI: 10.3389/fcimb.2022.855229
EFG1, Everyone's Favorite Gene in Candida albicans: A Comprehensive Literature Review
Abstract
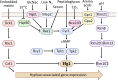
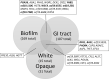
Candida sp. are among the most common fungal commensals found in the human microbiome. Although Candida can be found residing harmlessly on the surface of the skin and mucosal membranes, these opportunistic fungi have the potential to cause superficial skin, nail, and mucus membrane infections as well as life threatening systemic infections. Severity of infection is dependent on both fungal and host factors including the immune status of the host. Virulence factors associated with Candida sp. pathogenicity include adhesin proteins, degradative enzymes, phenotypic switching, and morphogenesis. A central transcriptional regulator of morphogenesis, the transcription factor Efg1 was first characterized in Candida albicans in 1997. Since then, EFG1 has been referenced in the Candida literature over three thousand times, with the number of citations growing daily. Arguably one of the most well studied genes in Candida albicans, EFG1 has been referenced in nearly all contexts of Candida biology from the development of novel therapeutics to white opaque switching, hyphae morphology to immunology. In the review that follows we will synthesize the research that has been performed on this extensively studied transcription factor and highlight several important unanswered questions.
Keywords: Candida albicans; EFG1; biofilm; fungal pathogen; transcription factor; transcription factor regulatory network; virulence.
Copyright © 2022 Glazier.
Conflict of interest statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Candida albicans Double Mutants Lacking both EFG1 and WOR1 Can Still Switch to Opaque.mSphere. 2020 Sep 23;5(5):e00918-20. doi: 10.1128/mSphere.00918-20. mSphere. 2020. PMID: 32968010 Free PMC article.
-
Collaboration between Antagonistic Cell Type Regulators Governs Natural Variation in the Candida albicans Biofilm and Hyphal Gene Expression Network.mBio. 2022 Oct 26;13(5):e0193722. doi: 10.1128/mbio.01937-22. Epub 2022 Aug 22. mBio. 2022. PMID: 35993746 Free PMC article.
-
Discovery of a "white-gray-opaque" tristable phenotypic switching system in candida albicans: roles of non-genetic diversity in host adaptation.PLoS Biol. 2014 Apr 1;12(4):e1001830. doi: 10.1371/journal.pbio.1001830. eCollection 2014 Apr. PLoS Biol. 2014. PMID: 24691005 Free PMC article.
-
Co-regulation of pathogenesis with dimorphism and phenotypic switching in Candida albicans, a commensal and a pathogen.Int J Med Microbiol. 2002 Oct;292(5-6):299-311. doi: 10.1078/1438-4221-00215. Int J Med Microbiol. 2002. PMID: 12452278 Review.
-
White-opaque switching in Candida albicans: cell biology, regulation, and function.Microbiol Mol Biol Rev. 2024 Jun 27;88(2):e0004322. doi: 10.1128/mmbr.00043-22. Epub 2024 Mar 28. Microbiol Mol Biol Rev. 2024. PMID: 38546228 Free PMC article. Review.
Cited by
-
Biosynthesis, characterization, and antifungal activity of plant-mediated silver nanoparticles using Cnidium monnieri fruit extract.Front Microbiol. 2023 Nov 20;14:1291030. doi: 10.3389/fmicb.2023.1291030. eCollection 2023. Front Microbiol. 2023. PMID: 38053552 Free PMC article.
-
Myricetin Exerts Antibiofilm Effects on Candida albicans by Targeting the RAS1/cAMP/EFG1 Pathway and Disruption of the Hyphal Network.J Fungi (Basel). 2025 May 21;11(5):398. doi: 10.3390/jof11050398. J Fungi (Basel). 2025. PMID: 40422732 Free PMC article.
-
Staphylococcus aureus wraps around Candida albicans and synergistically escapes from Neutrophil extracellular traps.Front Immunol. 2024 Jul 10;15:1422440. doi: 10.3389/fimmu.2024.1422440. eCollection 2024. Front Immunol. 2024. PMID: 39050841 Free PMC article.
-
Molecular association of Candida albicans and vulvovaginal candidiasis: focusing on a solution.Front Cell Infect Microbiol. 2023 Oct 13;13:1245808. doi: 10.3389/fcimb.2023.1245808. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 37900321 Free PMC article. Review.
-
The Endocytosis Adaptor Sla1 Facilitates Drug Susceptibility and Fungal Pathogenesis Through Sla1-Efg1 Regulating System in Candida albicans.Infect Drug Resist. 2024 Oct 22;17:4577-4588. doi: 10.2147/IDR.S483623. eCollection 2024. Infect Drug Resist. 2024. PMID: 39464835 Free PMC article.
References
-
- Bharucha N., Chabrier-Roselló Y., Xu T., Johnson C., Sobczynski S., Song Q., et al. . (2011). A Large-Scale Complex Haploinsufficiency-Based Genetic Interaction Screen in Candida Albicans: Analysis of the RAM Network During Morphogenesis. PloS Genet. 7 (4), e1002058. doi: 10.1371/journal.pgen.1002058 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

