Lactobacillus plantarum Disrupts S. mutans-C. albicans Cross-Kingdom Biofilms
- PMID: 35392605
- PMCID: PMC8980721
- DOI: 10.3389/fcimb.2022.872012
Lactobacillus plantarum Disrupts S. mutans-C. albicans Cross-Kingdom Biofilms
Abstract
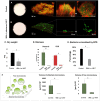
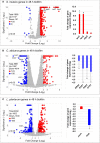
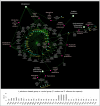
Dental caries, an ecological dysbiosis of oral microflora, initiates from the virulent biofilms formed on tooth surfaces where cariogenic microorganisms metabolize dietary carbohydrates, producing acid that demineralizes tooth enamel. Forming cariogenic biofilms, Streptococcus mutans and Candida albicans are well-recognized and emerging pathogens for dental caries. Recently, probiotics have demonstrated their potential in treating biofilm-related diseases, including caries. However, limited studies have assessed their effect on cariogenic bacteria-fungi cross-kingdom biofilm formation and their underlying interactions. Here, we assessed the effect of four probiotic Lactobacillus strains (Lactobacillus rhamnosus ATCC 2836, Lactobacillus plantarum ATCC 8014, Lactobacillus plantarum ATCC 14917, and Lactobacillus salivarius ATCC 11741) on S. mutans and C. albicans using a comprehensive multispecies biofilm model that mimicked high caries risk clinical conditions. Among the tested probiotic species, L. plantarum demonstrated superior inhibition on the growth of C. albicans and S. mutans, disruption of virulent biofilm formation with reduced bacteria and exopolysaccharide (EPS) components, and formation of virulent microcolonies structures. Transcriptome analysis (RNA sequencing) further revealed disruption of S. mutans and C. albicans cross-kingdom interactions with added L. plantarum. Genes of S. mutans and C. albicans involved in metabolic pathways (e.g., EPS formation, carbohydrate metabolism, glycan biosynthesis, and metabolism) were significantly downregulated. More significantly, genes related to C. albicans resistance to antifungal medication (ERG4), fungal cell wall chitin remodeling (CHT2), and resistance to oxidative stress (CAT1) were also significantly downregulated. In contrast, Lactobacillus genes plnD, plnG, and plnN that contribute to antimicrobial peptide plantaricin production were significantly upregulated. Our novel study findings support further assessment of the potential role of probiotic L. plantarum for cariogenic biofilm control.
Keywords: Candida albicans; Lactobacillus plantarum; Streptococcus mutans; cross-kingdom interactions; dental caries; multispecies biofilms.
Copyright © 2022 Zeng, Fadaak, Alomeir, Wu, Rustchenko, Qing, Bao, Gilbert and Xiao.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

