Chromosome-level genome assembly of grass carp (Ctenopharyngodon idella) provides insights into its genome evolution
- PMID: 35392810
- PMCID: PMC8988418
- DOI: 10.1186/s12864-022-08503-x
Chromosome-level genome assembly of grass carp (Ctenopharyngodon idella) provides insights into its genome evolution
Abstract
Background: The grass carp has great economic value and occupies an important evolutionary position. Genomic information regarding this species could help better understand its rapid growth rate as well as its unique body plan and environmental adaptation.
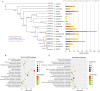
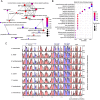
Results: We assembled the chromosome-level grass carp genome using the PacBio sequencing and chromosome structure capture technique. The final genome assembly has a total length of 893.2 Mb with a contig N50 of 19.3 Mb and a scaffold N50 of 35.7 Mb. About 99.85% of the assembled contigs were anchored into 24 chromosomes. Based on the prediction, this genome contained 30,342 protein-coding genes and 43.26% repetitive sequences. Furthermore, we determined that the large genome size can be attributed to the DNA-mediated transposable elements which accounted for 58.9% of the repetitive sequences in grass carp. We identified that the grass carp has only 24 pairs of chromosomes due to the fusion of two ancestral chromosomes. Enrichment analyses of significantly expanded and positively selected genes reflected evolutionary adaptation of grass carp to the feeding habits. We also detected the loss of conserved non-coding regulatory elements associated with the development of the immune system, nervous system, and digestive system, which may be critical for grass carp herbivorous traits.
Conclusions: The high-quality reference genome reported here provides a valuable resource for the genetic improvement and molecular-guided breeding of the grass carp.
Keywords: Adaptive evolution; Chromosome-level genome; Comparative genomics; Cyprinid fish; Grass carp.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



Similar articles
-
Comparative genomic analysis of immune-related genes and chemosensory receptors provides insights into the evolution and adaptation of four major domesticated Asian carps.BMC Genomics. 2025 May 26;26(1):529. doi: 10.1186/s12864-025-11719-2. BMC Genomics. 2025. PMID: 40419972 Free PMC article.
-
The draft genome of the grass carp (Ctenopharyngodon idellus) provides insights into its evolution and vegetarian adaptation.Nat Genet. 2015 Jun;47(6):625-31. doi: 10.1038/ng.3280. Epub 2015 May 4. Nat Genet. 2015. PMID: 25938946
-
The telomere-to-telomere gapless genome of grass carp provides insights for genetic improvement.Gigascience. 2025 Jan 6;14:giaf059. doi: 10.1093/gigascience/giaf059. Gigascience. 2025. PMID: 40531666 Free PMC article.
-
Chromosome-level genome assembly of the mud carp (Cirrhinus molitorella) using PacBio HiFi and Hi-C sequencing.Sci Data. 2024 Nov 19;11(1):1249. doi: 10.1038/s41597-024-04075-5. Sci Data. 2024. PMID: 39562583 Free PMC article.
-
High-quality chromosome-level genome assembly of Litsea coreana L. provides insights into Magnoliids evolution and flavonoid biosynthesis.Genomics. 2022 Jul;114(4):110394. doi: 10.1016/j.ygeno.2022.110394. Epub 2022 Jun 2. Genomics. 2022. PMID: 35659563 Review.
Cited by
-
Genetic improvement and genomic resources of important cyprinid species: status and future perspectives for sustainable production.Front Genet. 2024 Sep 19;15:1398084. doi: 10.3389/fgene.2024.1398084. eCollection 2024. Front Genet. 2024. PMID: 39364006 Free PMC article. Review.
-
Chromosome-Level Assembly of Male Opsariichthys bidens Genome Provides Insights into the Regulation of the GnRH Signaling Pathway and Genome Evolution.Biology (Basel). 2022 Oct 13;11(10):1500. doi: 10.3390/biology11101500. Biology (Basel). 2022. PMID: 36290404 Free PMC article.
-
Anti-infective immune functions of type IV interferon in grass carp ( Ctenopharyngodon idella): A novel antibacterial and antiviral interferon in lower vertebrates.Zool Res. 2024 Sep 18;45(5):972-982. doi: 10.24272/j.issn.2095-8137.2024.008. Zool Res. 2024. PMID: 39085753 Free PMC article.
-
A chromosome-level, haplotype-resolved genome assembly and annotation for the Eurasian minnow (Leuciscidae: Phoxinus phoxinus) provide evidence of haplotype diversity.Gigascience. 2025 Jan 6;14:giae116. doi: 10.1093/gigascience/giae116. Gigascience. 2025. PMID: 39877992 Free PMC article.
-
Animal board invited review: Widespread adoption of genetic technologies is key to sustainable expansion of global aquaculture.Animal. 2022 Oct;16(10):100642. doi: 10.1016/j.animal.2022.100642. Epub 2022 Sep 29. Animal. 2022. PMID: 36183431 Free PMC article. Review.
References
-
- Krynak KL, Oldfield RG, Dennis PM, Durkalec M, Weldon C. A novel field technique to assess ploidy in introduced grass carp (Ctenopharyngodon idella, Cyprinidae) Biol Invasions. 2015;17(7):1931–1939.
-
- Wang D, Wu FX. China fishery statistical yearbook. BeiJing: China Agriculture Press; 2021.
-
- Wang Y, Lu Y, Zhang Y, Ning Z, Li Y, Zhao Q, Lu H, Huang R, Xia X, Feng Q, et al. The draft genome of the grass carp (Ctenopharyngodon idellus) provides insights into its evolution and vegetarian adaptation. Nat Genet. 2015;47(6):625–631. - PubMed
-
- Shendure J, Ji H. Next-generation DNA sequencing. Nat Biotechnol. 2008;26(10):1135–1145. - PubMed
-
- Aravanis AM, Lee M, Klausner RD. Next-generation sequencing of circulating tumor DNA for early Cancer detection. Cell. 2017;168(4):571–574. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

