Gene conversion: a non-Mendelian process integral to meiotic recombination
- PMID: 35393552
- PMCID: PMC9273591
- DOI: 10.1038/s41437-022-00523-3
Gene conversion: a non-Mendelian process integral to meiotic recombination
Abstract
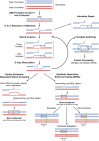
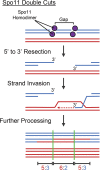
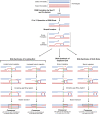
Meiosis is undoubtedly the mechanism that underpins Mendelian genetics. Meiosis is a specialised, reductional cell division which generates haploid gametes (reproductive cells) carrying a single chromosome complement from diploid progenitor cells harbouring two chromosome sets. Through this process, the hereditary material is shuffled and distributed into haploid gametes such that upon fertilisation, when two haploid gametes fuse, diploidy is restored in the zygote. During meiosis the transient physical connection of two homologous chromosomes (one originally inherited from each parent) each consisting of two sister chromatids and their subsequent segregation into four meiotic products (gametes), is what enables genetic marker assortment forming the core of Mendelian laws. The initiating events of meiotic recombination are DNA double-strand breaks (DSBs) which need to be repaired in a certain way to enable the homologous chromosomes to find each other. This is achieved by DSB ends searching for homologous repair templates and invading them. Ultimately, the repair of meiotic DSBs by homologous recombination physically connects homologous chromosomes through crossovers. These physical connections provided by crossovers enable faithful chromosome segregation. That being said, the DSB repair mechanism integral to meiotic recombination also produces genetic transmission distortions which manifest as postmeiotic segregation events and gene conversions. These processes are non-reciprocal genetic exchanges and thus non-Mendelian.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Allers T, Lichten M. Differential timing and control of noncrossover and crossover recombination during meiosis. Cell. 2001;106:47–57. - PubMed
-
- Bateson W. Mendel’s principles of heredity. Cambridge: Cambridge University Press; 1909.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

