Nascent alt-protein chemoproteomics reveals a pre-60S assembly checkpoint inhibitor
- PMID: 35393574
- PMCID: PMC9423127
- DOI: 10.1038/s41589-022-01003-9
Nascent alt-protein chemoproteomics reveals a pre-60S assembly checkpoint inhibitor
Abstract
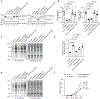
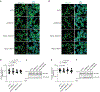
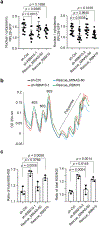
Many unannotated microproteins and alternative proteins (alt-proteins) are coencoded with canonical proteins, but few of their functions are known. Motivated by the hypothesis that alt-proteins undergoing regulated synthesis could play important cellular roles, we developed a chemoproteomic pipeline to identify nascent alt-proteins in human cells. We identified 22 actively translated alt-proteins or N-terminal extensions, one of which is post-transcriptionally upregulated by DNA damage stress. We further defined a nucleolar, cell-cycle-regulated alt-protein that negatively regulates assembly of the pre-60S ribosomal subunit (MINAS-60). Depletion of MINAS-60 increases the amount of cytoplasmic 60S ribosomal subunit, upregulating global protein synthesis and cell proliferation. Mechanistically, MINAS-60 represses the rate of late-stage pre-60S assembly and export to the cytoplasm. Together, these results implicate MINAS-60 as a potential checkpoint inhibitor of pre-60S assembly and demonstrate that chemoproteomics enables hypothesis generation for uncharacterized alt-proteins.
© 2022. The Author(s), under exclusive licence to Springer Nature America, Inc.
Figures



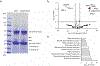









Comment in
-
Microproteins - lost in translation.Nat Chem Biol. 2022 Jun;18(6):581-582. doi: 10.1038/s41589-022-01007-5. Nat Chem Biol. 2022. PMID: 35393573 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

