NANOS3 suppresses premature spermatogonial differentiation to expand progenitors and fine-tunes spermatogenesis in mice
- PMID: 35394008
- PMCID: PMC9002807
- DOI: 10.1242/bio.059146
NANOS3 suppresses premature spermatogonial differentiation to expand progenitors and fine-tunes spermatogenesis in mice
Abstract
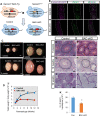
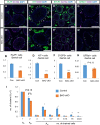
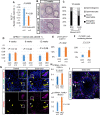
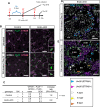
In the mouse testis, sperm originate from spermatogonial stem cells (SSCs). SSCs give rise to spermatogonial progenitors, which expand their population until entering the differentiation process that is precisely regulated by a fixed time-scaled program called the seminiferous cycle. Although this expansion process of progenitors is highly important, its regulatory mechanisms remain unclear. NANOS3 is an RNA-binding protein expressed in the progenitor population. We demonstrated that the conditional deletion of Nanos3 at a later embryonic stage results in the reduction of spermatogonial progenitors in the postnatal testis. This reduction was associated with the premature differentiation of progenitors. Furthermore, this premature differentiation caused seminiferous stage disagreement between adjacent spermatogenic cells, which influenced spermatogenic epithelial cycles, leading to disruption of the later differentiation pathway. Our study suggests that NANOS3 plays an important role in timing progenitor expansion to adjust to the proper differentiation timing by blocking the retinoic acid (RA) signaling pathway.
Keywords: Mouse; Nanos3; Retinoic acid; Spermatogenesis; Testis.
© 2022. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interests The authors declare no competing or financial interests.
Figures


Similar articles
-
Differential RA responsiveness among subsets of mouse late progenitor spermatogonia.Reproduction. 2021 May 5;161(6):645-655. doi: 10.1530/REP-21-0031. Reproduction. 2021. PMID: 33835049 Free PMC article.
-
Testicular Architecture Is Critical for Mediation of Retinoic Acid Responsiveness by Undifferentiated Spermatogonial Subtypes in the Mouse.Stem Cell Reports. 2018 Feb 13;10(2):538-552. doi: 10.1016/j.stemcr.2018.01.003. Epub 2018 Feb 1. Stem Cell Reports. 2018. PMID: 29398482 Free PMC article.
-
Stage-specific embryonic antigen 4 is a membrane marker for enrichment of porcine spermatogonial stem cells.Andrology. 2020 Nov;8(6):1923-1934. doi: 10.1111/andr.12870. Epub 2020 Aug 9. Andrology. 2020. PMID: 32691968
-
Epigenetic priming as a mechanism of predetermination of spermatogonial stem cell fate.Andrology. 2023 Jul;11(5):918-926. doi: 10.1111/andr.13332. Epub 2022 Nov 11. Andrology. 2023. PMID: 36333990 Review.
-
Spermatogonial stem cells and spermatogenesis in mice, monkeys and men.Stem Cell Res. 2018 May;29:207-214. doi: 10.1016/j.scr.2018.04.009. Epub 2018 Apr 21. Stem Cell Res. 2018. PMID: 29730571 Free PMC article. Review.
Cited by
-
Emerging Roles of NANOS RNA-Binding Proteins in Cancer.Int J Mol Sci. 2022 Aug 20;23(16):9408. doi: 10.3390/ijms23169408. Int J Mol Sci. 2022. PMID: 36012673 Free PMC article.
References
-
- Beedle, M. T., Stevison, F., Zhong, G., Topping, T., Hogarth, C., Isoherranen, N. and Griswold, M. D. (2019). Sources of all-trans retinal oxidation independent of the aldehyde dehydrogenase 1A isozymes exist in the postnatal testis. Biol. Reprod 100, 547-560. 10.1093/biolre/ioy200 - DOI - PMC - PubMed
-
- Endo, T., Romer, K. A., Anderson, E. L., Baltus, A. E., De Rooij, D. G. and Page, D. C. (2015). Periodic retinoic acid-STRA8 signaling intersects with periodic germ-cell competencies to regulate spermatogenesis. Proc. Natl. Acad. Sci. U. S. A 112, E2347-E2356. 10.1073/pnas.1505683112 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

