Maturation of hiPSC-derived cardiomyocytes promotes adult alternative splicing of SCN5A and reveals changes in sodium current associated with cardiac arrhythmia
- PMID: 35394010
- PMCID: PMC10022870
- DOI: 10.1093/cvr/cvac059
Maturation of hiPSC-derived cardiomyocytes promotes adult alternative splicing of SCN5A and reveals changes in sodium current associated with cardiac arrhythmia
Abstract
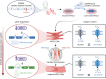
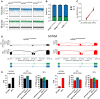
Aims: Human-induced pluripotent stem cell-cardiomyocytes (hiPSC-CMs) are widely used to study arrhythmia-associated mutations in ion channels. Among these, the cardiac sodium channel SCN5A undergoes foetal-to-adult isoform switching around birth. Conventional hiPSC-CM cultures, which are phenotypically foetal, have thus far been unable to capture mutations in adult gene isoforms. Here, we investigated whether tri-cellular cross-talk in a three-dimensional (3D) cardiac microtissue (MT) promoted post-natal SCN5A maturation in hiPSC-CMs.
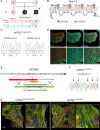
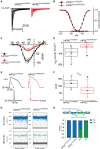
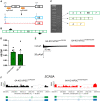
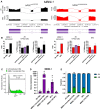
Methods and results: We derived patient hiPSC-CMs carrying compound mutations in the adult SCN5A exon 6B and exon 4. Electrophysiological properties of patient hiPSC-CMs in monolayer were not altered by the exon 6B mutation compared with isogenic controls since it is not expressed; further, CRISPR/Cas9-mediated excision of the foetal exon 6A did not promote adult SCN5A expression. However, when hiPSC-CMs were matured in 3D cardiac MTs, SCN5A underwent isoform switch and the functional consequences of the mutation located in exon 6B were revealed. Up-regulation of the splicing factor muscleblind-like protein 1 (MBNL1) drove SCN5A post-natal maturation in microtissues since its overexpression in hiPSC-CMs was sufficient to promote exon 6B inclusion, whilst knocking-out MBNL1 failed to foster isoform switch.
Conclusions: Our study shows that (i) the tri-cellular cardiac microtissues promote post-natal SCN5A isoform switch in hiPSC-CMs, (ii) adult splicing of SCN5A is driven by MBNL1 in these tissues, and (iii) this model can be used for examining post-natal cardiac arrhythmias due to mutations in the exon 6B.
Translational perspective: The cardiac sodium channel is essential for conducting the electrical impulse in the heart. Postnatal alternative splicing regulation causes mutual exclusive inclusion of fetal or adult exons of the corresponding gene, SCN5A. Typically, immature hiPSCCMs fall short in studying the effect of mutations located in the adult exon. We describe here that an innovative tri-cellular three-dimensional cardiac microtissue culture promotes hiPSC-CMs maturation through upregulation of MBNL1, thus revealing the effect of a pathogenic genetic variant located in the SCN5A adult exon. These results help advancing the use of hiPSC-CMs in studying adult heart disease and for developing personalized medicine applications.
Keywords: SCN5A; cardiac arrhythmias; cardiac microtissue; cardiac sodium channel; human-induced pluripotent stem cell-derived cardiomyocytes.
© The Author(s) 2022. Published by Oxford University Press on behalf of the European Society of Cardiology.
Conflict of interest statement
Conflict of interest: C.L.M. is co-founder of Ncardia bv.
Figures

References
-
- van den Berg CW, Elliott DA, Braam SR, Mummery CL, Davis RP. Differentiation of human pluripotent stem cells to cardiomyocytes under defined conditions. Methods Mol Biol 2016;1353:163–180. - PubMed
-
- Wilde AAM, Amin AS. Clinical spectrum of SCN5A mutations: long QT syndrome, Brugada syndrome, and cardiomyopathy. JACC Clin Electrophysiol 2018;4:569–579. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

