Epitope mapping of neutralising anti-SARS-CoV-2 monoclonal antibodies: Implications for immunotherapy and vaccine design
- PMID: 35394093
- PMCID: PMC9111153
- DOI: 10.1002/rmv.2347
Epitope mapping of neutralising anti-SARS-CoV-2 monoclonal antibodies: Implications for immunotherapy and vaccine design
Abstract
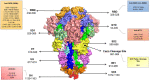
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the causative agent of the coronavirus disease 2019 (COVID-19) pandemic. This disease has currently affected more than 346 million people and resulted in more than 5.5 million deaths in many countries. Neutralising monoclonal antibodies (MAbs) against the SARS-CoV-2 virus could serve as prophylactic/therapeutic agents in COVID-19 infection by providing passive protection against the virus in individuals. Until now, no Food and Drug Administration/European Medicines Agency-approved neutralising MAb against SARS-CoV-2 virus exists in the market, though a number of MAbs have been authorised for emergency use. Therefore, there is an urgent need for development of efficient anti-SARS-CoV-2 neutralising MAbs for use in the clinic. Moreover, neutralising anti-SARS-CoV-2 MAbs could be used as beneficial tools for designing epitope-based vaccines against the virus. Given that the target epitope of a MAb is a crucial feature influencing its neutralising potency, target epitopes of neutralising anti-SARS-CoV-2 MAbs already reported in the literature and reactivity of these MAbs with SARS-CoV-2 variants are reviewed herein.
Keywords: COVID-19; RBD; SARS-CoV-2 virus; epitope mapping; immunotherapy; neutralising monoclonal antibody.
© 2022 John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- WHO. Coronavirus Disease (COVID‐19) Pandemic. https://www.who.int/emergencies/diseases/novel‐coronavirus‐2019
-
- Weekly Epidemiological Update on COVID‐19 ‐ 25 January 2022. https://www.who.int/publications/m/item/weekly‐epidemiological‐update‐on...
-
- Actemra EUA Letter of Authorization . https://www.fda.gov/news‐events/press‐announcements/coronavirus‐covid‐19...
-
- U.S. Food and Drug Administration . Center for Drug Evaluation and Research FDA Briefing Document Antimicrobial Drugs Advisory Committee Meeting November. 30; 2021. https://www.fda.gov/media/154418/download
-
- Paxlovid EUA Letter of Authorization . https://www.fda.gov/media/155049/download
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

