KRAS Mutants Upregulate Integrin β4 to Promote Invasion and Metastasis in Colorectal Cancer
- PMID: 35394541
- PMCID: PMC9357101
- DOI: 10.1158/1541-7786.MCR-21-0994
KRAS Mutants Upregulate Integrin β4 to Promote Invasion and Metastasis in Colorectal Cancer
Abstract
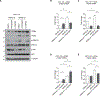
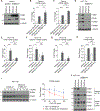
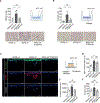
KRAS mutation in colorectal cancer is associated with aggressive tumor behavior through increased invasiveness and higher rates of lung metastases, but the biological mechanisms behind these features are not fully understood. In this study, we show that KRAS-mutant colorectal cancer upregulates integrin α6β4 through ERK/MEK signaling. Knocking-out integrin β4 (ITGB4) specifically depleted the expression of integrin α6β4 and this resulted in a reduction in the invasion and migration ability of the cancer cells. We also observed a reduction in the number and area of lung metastatic foci in mice that were injected with ITGB4 knockout KRAS-mutant colorectal cancer cells compared with the mice injected with ITGB4 wild-type KRAS-mutant colorectal cancer cells, while no difference was observed in liver metastases. Inhibiting integrin α6β4 in KRAS-mutant colorectal cancer could be a potential therapeutic target to diminish the KRAS-invasive phenotype and associated pulmonary metastasis rate.
Implications: Knocking-out ITGB4, which is overexpressed in KRAS-mutant colorectal cancer and promotes tumor aggressiveness, diminishes local invasiveness and rates of pulmonary metastasis.
©2022 American Association for Cancer Research.
Conflict of interest statement
Disclosure of Potential Conflicts of Interest:
JGA has received honorarium for being a consultant with the following: Medtronics, Ethicon J&J, Da Vinci Intuitive Surgical. JJS has received travel support from Intuitive Surgical Inc. for fellow education (2015) and has served as a clinical advisor for Guardant Health, Inc (2019). The other co-authors have no conflicts of interest to disclose.
Figures




References
-
- Hynes RO. Integrins: bidirectional, allosteric signaling machines. Cell. 2002;110:673–87. - PubMed
-
- Giancotti FG, Ruoslahti E. Integrin signaling. Science. 1999;285:1028–32. - PubMed
-
- Lipscomb EA, Mercurio AM. Mobilization and activation of a signaling competent alpha6beta4integrin underlies its contribution to carcinoma progression. Cancer Metastasis Rev. 2005;24:413–23. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

