Cytotoxic granzyme C-expressing ILC1s contribute to antitumor immunity and neonatal autoimmunity
- PMID: 35394814
- PMCID: PMC9233921
- DOI: 10.1126/sciimmunol.abi8642
Cytotoxic granzyme C-expressing ILC1s contribute to antitumor immunity and neonatal autoimmunity
Abstract
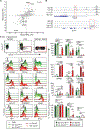
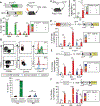
Innate lymphocytes are integral components of the cellular immune system that can coordinate host defense against a multitude of challenges and trigger immunopathology when dysregulated. Natural killer (NK) cells and innate lymphoid cells (ILCs) are innate immune effectors postulated to functionally mirror conventional cytotoxic T lymphocytes and helper T cells, respectively. Here, we showed that the cytolytic molecule granzyme C was expressed in cells with the phenotype of type 1 ILCs (ILC1s) in mouse liver and salivary gland. Cell fate-mapping and transfer studies revealed that granzyme C-expressing innate lymphocytes could be derived from ILC progenitors and did not interconvert with NK cells, ILC2s, or ILC3s. Granzyme C defined a maturation state of ILC1s. These granzyme C-expressing ILC1s required the transcription factors T-bet and, to a lesser extent, Eomes and support from transforming growth factor-β (TGF-β) signaling for their maintenance in the salivary gland. In a transgenic mouse breast cancer model, depleting ILC1s caused accelerated tumor growth. ILC1s gained granzyme C expression following interleukin-15 (IL-15) stimulation, which enabled perforin-mediated cytotoxicity. Constitutive activation of STAT5, a transcription factor regulated by IL-15, in granzyme C-expressing ILC1s triggered lethal perforin-dependent autoimmunity in neonatal mice. Thus, granzyme C marks a cytotoxic effector state of ILC1s, broadening their function beyond "helper-like" lymphocytes.
Conflict of interest statement
Competing Interests:
MSKCC has filed a patent application with the U.S. Patent and Trademark Office directed toward targeting ILC1 IL-15 signaling for cancer immunotherapy. MOL is an SAB member of and holds equity or stock options in Amberstone Biosciences Inc. AYR is an SAB member of and holds equity or stock options in Sonoma Biotherapeutic, Surface Oncology, RAPT Therapeutics, and Vedanta Biosciences. He is an SAB member of BioInvent. A.Y.R. holds IP rights for therapeutic Treg cell depleting antibodies licensed to Takeda that is unrelated to the content of the present study.
Figures






Comment in
-
ILC killer: Qu'est-ce que c'est?Sci Immunol. 2022 Apr 8;7(70):eabo2159. doi: 10.1126/sciimmunol.abo2159. Epub 2022 Apr 8. Sci Immunol. 2022. PMID: 35394813
References
-
- Vivier E, Artis D, Colonna M, Diefenbach A, Di Santo JP, Eberl G, Koyasu S, Locksley RM, McKenzie ANJ, Mebius RE, Powrie F, Spits H, Innate Lymphoid Cells: 10 Years On. Cell 174, 1054–1066 (2018). - PubMed
-
- Friedrich C, Taggenbrock R, Doucet-Ladeveze R, Golda G, Moenius R, Arampatzi P, Kragten NAM, Kreymborg K, Gomez de Aguero M, Kastenmuller W, Saliba AE, Grun D, van Gisbergen K, Gasteiger G, Effector differentiation downstream of lineage commitment in ILC1s is driven by Hobit across tissues. Nat Immunol 22, 1256–1267 (2021). - PMC - PubMed
-
- Di Censo C, Marotel M, Mattiola I, Muller L, Scarno G, Pietropaolo G, Peruzzi G, Laffranchi M, Mazej J, Hasim MS, Asif S, Russo E, Tomaipitinca L, Stabile H, Lee SH, Vian L, Gadina M, Gismondi A, Shih HY, Mikami Y, Capuano C, Bernardini G, Bonelli M, Sozzani S, Diefenbach A, Ardolino M, Santoni A, Sciume G, Granzyme A and CD160 expression delineates ILC1 with graded functions in the mouse liver. Eur J Immunol 51, 2568–2575 (2021). - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

