Human enteric viruses autonomously shape inflammatory bowel disease phenotype through divergent innate immunomodulation
- PMID: 35394816
- PMCID: PMC9416881
- DOI: 10.1126/sciimmunol.abn6660
Human enteric viruses autonomously shape inflammatory bowel disease phenotype through divergent innate immunomodulation
Abstract
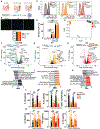
Altered enteric microorganisms in concert with host genetics shape inflammatory bowel disease (IBD) phenotypes. However, insight is limited to bacteria and fungi. We found that eukaryotic viruses and bacteriophages (collectively, the virome), enriched from non-IBD, noninflamed human colon resections, actively elicited atypical anti-inflammatory innate immune programs. Conversely, ulcerative colitis or Crohn's disease colon resection viromes provoked inflammation, which was successfully dampened by non-IBD viromes. The IBD colon tissue virome was perturbed, including an increase in the enterovirus B species of eukaryotic picornaviruses, not previously detected in fecal virome studies. Mice humanized with non-IBD colon tissue viromes were protected from intestinal inflammation, whereas IBD virome mice exhibited exacerbated inflammation in a nucleic acid sensing-dependent fashion. Furthermore, there were detrimental consequences for IBD patient-derived intestinal epithelial cells bearing loss-of-function mutations within virus sensor MDA5 when exposed to viromes. Our results demonstrate that innate recognition of IBD or non-IBD human viromes autonomously influences intestinal homeostasis and disease phenotypes. Thus, perturbations in the intestinal virome, or an altered ability to sense the virome due to genetic variation, contribute to the induction of IBD. Harnessing the virome may offer therapeutic and biomarker potential.
Conflict of interest statement
Figures


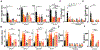


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

