Pten heterozygosity restores neuronal morphology in fragile X syndrome mice
- PMID: 35394871
- PMCID: PMC9169627
- DOI: 10.1073/pnas.2109448119
Pten heterozygosity restores neuronal morphology in fragile X syndrome mice
Abstract
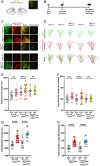
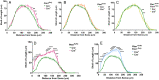
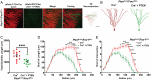
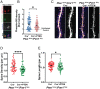
Genetic studies of hippocampal granule neuron development have been used to elucidate cellular functions of Pten and Fmr1. While mutations in each gene cause neurodevelopmental disorders such as autism and fragile X syndrome, how Pten and Fmr1 function alone or together during normal development is not known. Moreover, Pten mRNA is bound by the fragile X mental retardation protein (FMRP) RNA binding protein, but how this physical interaction impinges on phosphatase and tensin homolog protein (PTEN) expression is not known. To understand the interaction of PTEN and FMRP, we investigated the dentate gyrus granule neuron development in Pten and Fmr1 knockout (KO) mice. Interestingly, heterozygosity of Pten restored Fmr1 KO cellular phenotypes, including dendritic arborization, and spine density, while PTEN protein expression was significantly increased in Fmr1 KO animals. However, complete deletion of both Pten and Fmr1 resulted in a dramatic increase in dendritic length, spine density, and spine length. In addition, overexpression of PTEN in Fmr1 KO Pten heterozygous background reduced dendritic length, arborization, spine density, and spine length including pS6 levels. Our findings suggest that PTEN levels are negatively regulated by FMRP, and some Fmr1 KO phenotypes are caused by dysregulation of PTEN protein. These observations provide evidence for the genetic interaction of PTEN and FMRP and a possible mechanistic basis for the pathogenesis of Fmr1-related fragile X neurodevelopmental disorders.
Keywords: Fmr1; Pten; arborization; dentate gyrus; spine density.
Conflict of interest statement
The authors declare no competing interest.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

