Extracellular vesicle proteomic analysis leads to the discovery of HDGF as a new factor in multiple myeloma biology
- PMID: 35395072
- PMCID: PMC9198912
- DOI: 10.1182/bloodadvances.2021006187
Extracellular vesicle proteomic analysis leads to the discovery of HDGF as a new factor in multiple myeloma biology
Abstract
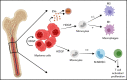
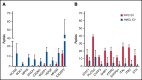
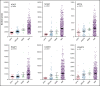
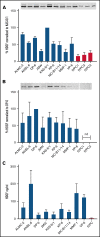
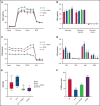
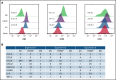
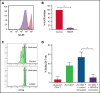
Identifying factors secreted by multiple myeloma (MM) cells that may contribute to MM tumor biology and progression is of the utmost importance. In this study, hepatoma-derived growth factor (HDGF) was identified as a protein present in extracellular vesicles (EVs) released from human MM cell lines (HMCLs). Investigation of the role of HDGF in MM cell biology revealed lower proliferation of HMCLs following HDGF knockdown and AKT phosphorylation following the addition of exogenous HDGF. Metabolic analysis demonstrated that HDGF enhances the already high glycolytic levels of HMCLs and significantly lowers mitochondrial respiration, indicating that HDGF may play a role in myeloma cell survival and/or act in a paracrine manner on cells in the bone marrow (BM) tumor microenvironment (ME). Indeed, HDGF polarizes macrophages to an M1-like phenotype and phenotypically alters naïve CD14+ monocytes to resemble myeloid-derived suppressor cells which are functionally suppressive. In summary, HDGF is a novel factor in MM biology and may function to both maintain MM cell viability as well as modify the tumor ME.
© 2022 by The American Society of Hematology. Licensed under Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International (CC BY-NC-ND 4.0), permitting only noncommercial, nonderivative use with attribution. All other rights reserved.
Figures

References
-
- van Niel G, D’Angelo G, Raposo G. Shedding light on the cell biology of extracellular vesicles. Nat Rev Mol Cell Biol. 2018;19(4):213-228. - PubMed
-
- Moloudizargari M, Abdollahi M, Asghari MH, Zimta AA, Neagoe IB, Nabavi SM. The emerging role of exosomes in multiple myeloma. Blood Rev. 2019;38:100595. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

