STRipy: A graphical application for enhanced genotyping of pathogenic short tandem repeats in sequencing data
- PMID: 35395114
- PMCID: PMC9541159
- DOI: 10.1002/humu.24382
STRipy: A graphical application for enhanced genotyping of pathogenic short tandem repeats in sequencing data
Abstract
Expansions of short tandem repeats (STRs) have been implicated as the causal variant in over 50 diseases known to date. There are several tools which can genotype STRs from high-throughput sequencing (HTS) data. However, running these tools out of the box only allows around half of the known disease-causing loci to be genotyped. Furthermore, the genotypes estimated at these loci are often underestimated with maximum lengths limited to either the read or fragment length, which is less than the pathogenic cutoff for some diseases. Although analysis tools can be customized to genotype extra loci, this requires proficiency in bioinformatics to set up, limiting their widespread usage by other researchers and clinicians. To address these issues, we have developed a new software called STRipy, which is able to target all known disease-causing STRs from HTS data. We created an intuitive graphical interface for STRipy and significantly simplified the detection of STRs expansions. Moreover, we genotyped all disease loci for over two and half thousand samples to provide population-wide distributions to assist with interpretation of results. We believe the simplicity and breadth of STRipy will increase the genotyping of STRs in sequencing data resulting in further diagnoses of rare STR diseases.
Keywords: bioinformatics tools; high-throughput sequencing; pathogenic mutations; rare diseases; short tandem repeats.
© 2022 The Authors. Human Mutation published by Wiley Periodicals LLC.
Conflict of interest statement
Egor Dolzhenko is an employee of Illumina, Inc., a public company that develops and markets systems for genetic analysis.
Figures
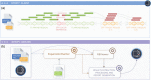

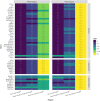
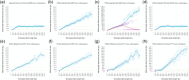
References
-
- Auton, A. , Abecasis, G. R. , Altshuler, D. M. , Durbin, R. M. , Bentley, D. R. , Chakravarti, A. , Clark, A. G. , Donnelly, P. , Eichler, E. E. , Flicek, P. , Gabriel, S. B. , Gibbs, R. A. , Green, E. D. , Hurles, M. E. , Knoppers, B. M. , Korbel, J. O. , Lander, E. S. , Lee, C. , Lehrach, H. , … Yao, L . (2015). A global reference for human genetic variation. Nature, 526(7571), 68–74. 10.1038/nature15393 - DOI - PMC - PubMed
-
- Corbett, M. A. , Kroes, T. , Veneziano, L. , Bennett, M. F. , Florian, R. , Schneider, A. L. , Coppola, A. , Licchetta, L. , Franceschetti, S. , Suppa, A. , Wenger, A. , Mei, D. , Pendziwiat, M. , Kaya, S. , Delledonne, M. , Straussberg, R. , Xumerle, L. , Regan, B. , Crompton, D. , … Gecz, J . (2019). Intronic ATTTC repeat expansions in STARD7 in familial adult myoclonic epilepsy linked to chromosome 2. Nature Communications, 10(1), 1–10. 10.1038/s41467-019-12671-y - DOI - PMC - PubMed
-
- Cortese, A. , Simone, R. , Sullivan, R. , Vandrovcova, J. , Tariq, H. , Yau, W. Y. , Humphrey, J. , Jaunmuktane, Z. , Sivakumar, P. , Polke, J. , Ilyas, M. , Tribollet, E. , Tomaselli, P. J. , Devigili, G. , Callegari, I. , Versino, M. , Salpietro, V. , Efthymiou, S. , Kaski, D. , … Houlden, H . (2019). Biallelic expansion of an intronic repeat in RFC1 is a common cause of late‐onset ataxia. Nature Genetics, 51(4), 649–658. 10.1038/s41588-019-0372-4 - DOI - PMC - PubMed
-
- Dashnow, H. , Lek, M. , Phipson, B. , Halman, A. , Sadedin, S. , Lonsdale, A. , Davis, M. , Lamont, P. , Clayton, J. S. , Laing, N. G. , MacArthur, D. G. , & Oshlack, A. (2018). STRetch: Detecting and discovering pathogenic short tandem repeat expansions. Genome Biology, 19(1), 1–13. 10.1186/s13059-018-1505-2 - DOI - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

