SETDB1 interactions with PELP1 contributes to breast cancer endocrine therapy resistance
- PMID: 35395812
- PMCID: PMC8991965
- DOI: 10.1186/s13058-022-01520-4
SETDB1 interactions with PELP1 contributes to breast cancer endocrine therapy resistance
Abstract
Background: Methyltransferase SETDB1 is highly expressed in breast cancer (BC), however, the mechanisms by which SETDB1 promotes BC progression to endocrine therapy resistance remains elusive. In this study, we examined the mechanisms by which SETDB1 contribute to BC endocrine therapy resistance.
Methods: We utilized therapy sensitive (MCF7 and ZR75), therapy resistant (MCF7-TamR, MCF7-FR, MCF7-PELP1cyto, MCF7-SETDB1) estrogen receptor alpha positive (ER+)BC models and conducted in vitro cell viability, colony formation, 3-dimensional cell growth assays to investigate the role of SETDB1 in endocrine resistance. RNA-seq of parental and SETDB1 knock down ER+ BC cells was used to identify unique pathways. SETDB1 interaction with PELP1 was identified by yeast-two hybrid screen and confirmed by immunoprecipitation and GST-pull down assays. Mechanistic studies were conducted using Western blotting, reporter gene assays, RT-qPCR, and in vitro methylation assays. Xenograft assays were used to establish the role of PELP1 in SETDB1 mediated BC progression.
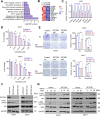
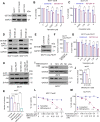
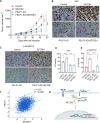
Results: RNA-seq analyses showed that SETDB1 regulates expression of a subset of estrogen receptor (ER) and Akt target genes that contribute to endocrine therapy resistance. Importantly, using yeast-two hybrid screen, we identified ER coregulator PELP1 as a novel interacting protein of SETDB1. Biochemical analyses confirmed SETDB1 and PELP1 interactions in multiple BC cells. Mechanistic studies confirmed that PELP1 is necessary for SETDB1 mediated Akt methylation and phosphorylation. Further, SETDB1 overexpression promotes tamoxifen resistance in BC cells, and PELP1 knockdown abolished these effects. Using xenograft model, we provided genetic evidence that PELP1 is essential for SETDB1 mediated BC progression in vivo. Analyses of TCGA datasets revealed SETDB1 expression is positively correlated with PELP1 expression in ER+ BC patients.
Conclusions: This study suggests that the PELP1/SETDB1 axis play an important role in aberrant Akt activation and serves as a novel target for treating endocrine therapy resistance in breast cancer.
Keywords: Akt; Breast cancer; PELP1; SETDB1; Therapy resistance.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing financial interests.
Figures




Similar articles
-
Interaction of transcription factor AP-2 gamma with proto-oncogene PELP1 promotes tumorigenesis by enhancing RET signaling.Mol Oncol. 2021 Apr;15(4):1146-1161. doi: 10.1002/1878-0261.12871. Epub 2021 Feb 9. Mol Oncol. 2021. PMID: 33269540 Free PMC article.
-
Inhibition of mTOR signaling reduces PELP1-mediated tumor growth and therapy resistance.Mol Cancer Ther. 2014 Jun;13(6):1578-88. doi: 10.1158/1535-7163.MCT-13-0877. Epub 2014 Mar 31. Mol Cancer Ther. 2014. PMID: 24688046 Free PMC article.
-
Significance of ER-Src axis in hormonal therapy resistance.Breast Cancer Res Treat. 2011 Nov;130(2):377-85. doi: 10.1007/s10549-010-1312-2. Epub 2010 Dec 24. Breast Cancer Res Treat. 2011. PMID: 21184269 Free PMC article.
-
PELP1: Structure, biological function and clinical significance.Gene. 2016 Jul 1;585(1):128-134. doi: 10.1016/j.gene.2016.03.017. Epub 2016 Mar 18. Gene. 2016. PMID: 26997260 Free PMC article. Review.
-
PELP1: A novel therapeutic target for hormonal cancers.IUBMB Life. 2010 Mar;62(3):162-9. doi: 10.1002/iub.287. IUBMB Life. 2010. PMID: 20014005 Free PMC article. Review.
Cited by
-
Hepatocellular SETDB1 Regulates Hepatic Ischemia-Reperfusion Injury through Targeting Lysine Methylation of ASK1 Signal.Research (Wash D C). 2023 Oct 31;6:0256. doi: 10.34133/research.0256. eCollection 2023. Research (Wash D C). 2023. PMID: 37915765 Free PMC article.
-
SETDB1 promotes progression through upregulation of SF3B4 expression and regulates the immunity in ovarian cancer.J Ovarian Res. 2024 Feb 5;17(1):34. doi: 10.1186/s13048-024-01358-8. J Ovarian Res. 2024. PMID: 38317200 Free PMC article.
-
Crosstalk of methylation and tamoxifen in breast cancer (Review).Mol Med Rep. 2024 Oct;30(4):180. doi: 10.3892/mmr.2024.13304. Epub 2024 Aug 12. Mol Med Rep. 2024. PMID: 39129315 Free PMC article. Review.
-
Anti-cancer targets and molecular mechanisms of formononetin in treating osteosarcoma based on network pharmacology.Aging (Albany NY). 2023 Oct 20;15(20):11489-11507. doi: 10.18632/aging.205139. Epub 2023 Oct 20. Aging (Albany NY). 2023. PMID: 37870753 Free PMC article.
-
Potential therapies for non-coding RNAs in breast cancer.Front Oncol. 2024 Sep 20;14:1452666. doi: 10.3389/fonc.2024.1452666. eCollection 2024. Front Oncol. 2024. PMID: 39372872 Free PMC article. Review.
References
-
- Turner NC, Neven P, Loibl S, Andre F. Advances in the treatment of advanced oestrogen-receptor-positive breast cancer. Lancet (London, England) 2017;389(10087):2403–2414. - PubMed
-
- Musgrove EA, Sutherland RL. Biological determinants of endocrine resistance in breast cancer. Nat Rev Cancer. 2009;9(9):631–643. - PubMed
-
- Viedma-Rodriguez R, Baiza-Gutman L, Salamanca-Gomez F, Diaz-Zaragoza M, Martinez-Hernandez G, Ruiz Esparza-Garrido R, Velazquez-Flores MA, Arenas-Aranda D. Mechanisms associated with resistance to tamoxifen in estrogen receptor-positive breast cancer (review) Oncol Rep. 2014;32(1):3–15. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

