Best practices for the interpretation and reporting of clinical whole genome sequencing
- PMID: 35395838
- PMCID: PMC8993917
- DOI: 10.1038/s41525-022-00295-z
Best practices for the interpretation and reporting of clinical whole genome sequencing
Abstract
Whole genome sequencing (WGS) shows promise as a first-tier diagnostic test for patients with rare genetic disorders. However, standards addressing the definition and deployment practice of a best-in-class test are lacking. To address these gaps, the Medical Genome Initiative, a consortium of leading health care and research organizations in the US and Canada, was formed to expand access to high quality clinical WGS by convening experts and publishing best practices. Here, we present best practice recommendations for the interpretation and reporting of clinical diagnostic WGS, including discussion of challenges and emerging approaches that will be critical to harness the full potential of this comprehensive test.
© 2022. The Author(s).
Conflict of interest statement
D.L.P. and R.J.T. are current employees and shareholders of Illumina Inc. The remaining authors declare no competing interests.
Figures
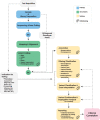
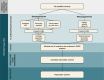
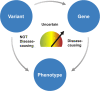
References
Publication types
LinkOut - more resources
Full Text Sources

