Cohort design and natural language processing to reduce bias in electronic health records research
- PMID: 35396454
- PMCID: PMC8993873
- DOI: 10.1038/s41746-022-00590-0
Cohort design and natural language processing to reduce bias in electronic health records research
Abstract
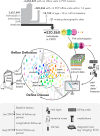
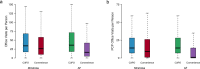
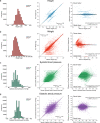
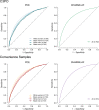
Electronic health record (EHR) datasets are statistically powerful but are subject to ascertainment bias and missingness. Using the Mass General Brigham multi-institutional EHR, we approximated a community-based cohort by sampling patients receiving longitudinal primary care between 2001-2018 (Community Care Cohort Project [C3PO], n = 520,868). We utilized natural language processing (NLP) to recover vital signs from unstructured notes. We assessed the validity of C3PO by deploying established risk models for myocardial infarction/stroke and atrial fibrillation. We then compared C3PO to Convenience Samples including all individuals from the same EHR with complete data, but without a longitudinal primary care requirement. NLP reduced the missingness of vital signs by 31%. NLP-recovered vital signs were highly correlated with values derived from structured fields (Pearson r range 0.95-0.99). Atrial fibrillation and myocardial infarction/stroke incidence were lower and risk models were better calibrated in C3PO as opposed to the Convenience Samples (calibration error range for myocardial infarction/stroke: 0.012-0.030 in C3PO vs. 0.028-0.046 in Convenience Samples; calibration error for atrial fibrillation 0.028 in C3PO vs. 0.036 in Convenience Samples). Sampling patients receiving regular primary care and using NLP to recover missing data may reduce bias and maximize generalizability of EHR research.
© 2022. The Author(s).
Conflict of interest statement
The authors declare the following financial and non-financial interests: A.A.P. receives sponsored research support from Bayer AG, IBM, Intel, and Verily. He has also received consulted fees from Novartis and Rakuten. He is a Venture Partner at GV and is compensated for this work. J.E.H. receives sponsored research support from Bayer AG and Gilead Sciences. J.E.H. has received research supplies from EcoNugenics. S.F.F. receives sponsored research support from Bayer AG and IBM. C.D.A. receives sponsored research support from Bayer AG and has consulted for ApoPharma and Invitae. P.B. receives sponsored research support from Bayer AG and IBM, and consults for Novartis. S.A.L. receives sponsored research support from Bristol Myers Squibb/Pfizer, Bayer AG, Boehringer Ingelheim, and Fitbit, has consulted for Bristol Myers Squibb/Pfizer and Bayer AG, and participates in a research collaboration with IBM. P.T.E. receives sponsored research support from Bayer AG and IBM Research and he has consulted for Bayer AG, Novartis, MyoKardia, and Quest Diagnostics. S.J.A. receives sponsored research support from Bristol Myers Squibb/Pfizer and has consulted for Bristol Myers Squibb/Pfizer and Fitbit. J.M.A. has received sponsored research support from Bristol Myers Squibb/Pfizer. C.D., J.M., H.M.E., A.D., and M.G. are employees of Bayer AG. The remaining authors declare no competing interests.
Figures


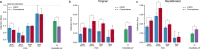

References
Grants and funding
- T32 HL094301/HL/NHLBI NIH HHS/United States
- R01 HL139731/HL/NHLBI NIH HHS/United States
- R01 HL092577/HL/NHLBI NIH HHS/United States
- R38 HL150212/HL/NHLBI NIH HHS/United States
- 18SFRN34110082/AHA/American Heart Association-American Stroke Association/United States
- R01 HL128914/HL/NHLBI NIH HHS/United States
- T32 HL007208/HL/NHLBI NIH HHS/United States
- R01 NS103924/NS/NINDS NIH HHS/United States
- 18SFRN34250007/AHA/American Heart Association-American Stroke Association/United States
- K01 HL148506/HL/NHLBI NIH HHS/United States
- K23 HL159243/HL/NHLBI NIH HHS/United States
- K24 HL105780/HL/NHLBI NIH HHS/United States
- R01 HL140224/HL/NHLBI NIH HHS/United States
- K24 HL153669/HL/NHLBI NIH HHS/United States
- R01 HL134893/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

