MAT2A facilitates PDCD6 methylation and promotes cell growth under glucose deprivation in cervical cancer
- PMID: 35396512
- PMCID: PMC8993843
- DOI: 10.1038/s41420-022-00987-6
MAT2A facilitates PDCD6 methylation and promotes cell growth under glucose deprivation in cervical cancer
Abstract
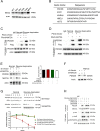
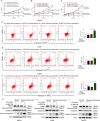
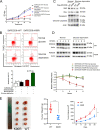
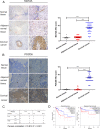
The underlying mechanisms of methionine adenosyltransferase 2 A (MAT2A)-mediated cervical cancer progression under nutrient stress are largely elusive. Therefore, our study aims to investigate molecular mechanism by which MAT2A-indcued cervical oncogenesis. The interaction between MAT2A and programmed cell death protein 6 (PDCD6) in cervical cancer cell lines was detected by immunoprecipitation, immunoblotting and mass spectrometric analysis. A panel of inhibitors that are linked to stress responsive kinases were utilized to detect related pathways by immunoblotting. Cell proliferation and apoptosis were investigated by CCK-8 and flow cytometry. Apoptosis related protein level of Bcl-2, Bax and Caspase-3 was also analyzed in cells with PDCD6 K90 methylation mutation. The association between MAT2A and PDCD6 was detected by immunohistochemistry and clinicopathological characteristics were further analyzed. We found that the interaction between MAT2A and PDCD6 is mediated by AMPK activation and facilitates PDCD6 K90 methylation and further promotes protein stability of PDCD6. Physiologically, expression of PDCD6 K90R leads to increased apoptosis and thus suppresses growth of cervical cancer cells under glucose deprivation. Furthermore, the clinical analysis indicates that the MAT2A protein level is positively associated with the PDCD6 level, and the high level of PDCD6 significantly correlates with poor prognosis and advanced stages of cervical cancer patients. We conclude that MAT2A facilitates PDCD6 methylation to promote cervical cancer growth under glucose deprivation, suggesting the regulatory role of MAT2A in cellular response to nutrient stress and cervical cancer progression.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

