Integrated analysis of expression profiles with meat quality traits in cattle
- PMID: 35396568
- PMCID: PMC8993808
- DOI: 10.1038/s41598-022-09998-w
Integrated analysis of expression profiles with meat quality traits in cattle
Abstract
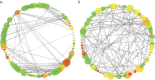
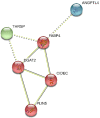
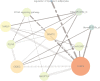
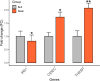
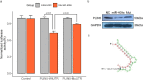
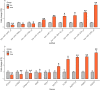
MicroRNAs (miRNAs) play a vital role in improving meat quality by binding to messenger RNAs (mRNAs). We performed an integrated analysis of miRNA and mRNA expression profiling between bulls and steers based on the differences in meat quality traits. Fat and fatty acids are the major phenotypic indices of meat quality traits to estimate between-group variance. In the present study, 90 differentially expressed mRNAs (DEGs) and 18 differentially expressed miRNAs (DEMs) were identified. Eighty-three potential DEG targets and 18 DEMs were used to structure a negative interaction network, and 75 matching target genes were shown in this network. Twenty-six target genes were designated as intersection genes, screened from 18 DEMs, and overlapped with the DEGs. Seventeen of these genes enriched to 19 terms involved in lipid metabolism. Subsequently, 13 DEGs and nine DEMs were validated using quantitative real-time PCR, and seven critical genes were selected to explore the influence of fat and fatty acids through hub genes and predict functional association. A dual-luciferase reporter and Western blot assays confirmed a predicted miRNA target (bta-miR-409a and PLIN5). These findings provide substantial evidence for molecular genetic controls and interaction among genes in cattle.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

