γ-Secretase in Alzheimer's disease
- PMID: 35396575
- PMCID: PMC9076685
- DOI: 10.1038/s12276-022-00754-8
γ-Secretase in Alzheimer's disease
Abstract
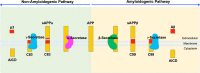
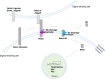
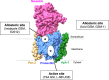
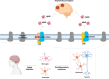
Alzheimer's disease (AD) is caused by synaptic and neuronal loss in the brain. One of the characteristic hallmarks of AD is senile plaques containing amyloid β-peptide (Aβ). Aβ is produced from amyloid precursor protein (APP) by sequential proteolytic cleavages by β-secretase and γ-secretase, and the polymerization of Aβ into amyloid plaques is thought to be a key pathogenic event in AD. Since γ-secretase mediates the final cleavage that liberates Aβ, γ-secretase has been widely studied as a potential drug target for the treatment of AD. γ-Secretase is a transmembrane protein complex containing presenilin, nicastrin, Aph-1, and Pen-2, which are sufficient for γ-secretase activity. γ-Secretase cleaves >140 substrates, including APP and Notch. Previously, γ-secretase inhibitors (GSIs) were shown to cause side effects in clinical trials due to the inhibition of Notch signaling. Therefore, more specific regulation or modulation of γ-secretase is needed. In recent years, γ-secretase modulators (GSMs) have been developed. To modulate γ-secretase and to understand its complex biology, finding the binding sites of GSIs and GSMs on γ-secretase as well as identifying transiently binding γ-secretase modulatory proteins have been of great interest. In this review, decades of findings on γ-secretase in AD are discussed.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

