Neither cardiac mitochondrial DNA variation nor copy number contribute to congenital heart disease risk
- PMID: 35397206
- PMCID: PMC9118105
- DOI: 10.1016/j.ajhg.2022.03.011
Neither cardiac mitochondrial DNA variation nor copy number contribute to congenital heart disease risk
Abstract
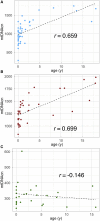
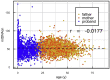
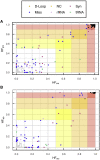
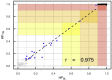
The well-established manifestation of mitochondrial mutations in functional cardiac disease (e.g., mitochondrial cardiomyopathy) prompted the hypothesis that mitochondrial DNA (mtDNA) sequence and/or copy number (mtDNAcn) variation contribute to cardiac defects in congenital heart disease (CHD). MtDNAcns were calculated and rare, non-synonymous mtDNA mutations were identified in 1,837 CHD-affected proband-parent trios, 116 CHD-affected singletons, and 114 paired cardiovascular tissue/blood samples. The variant allele fraction (VAF) of heteroplasmic variants in mitochondrial RNA from 257 CHD cardiovascular tissue samples was also calculated. On average, mtDNA from blood had 0.14 rare variants and 52.9 mtDNA copies per nuclear genome per proband. No variation with parental age at proband birth or CHD-affected proband age was seen. mtDNAcns in valve/vessel tissue (320 ± 70) were lower than in atrial tissue (1,080 ± 320, p = 6.8E-21), which were lower than in ventricle tissue (1,340 ± 280, p = 1.4E-4). The frequency of rare variants in CHD-affected individual DNA was indistinguishable from the frequency in an unaffected cohort, and proband mtDNAcns did not vary from those of CHD cohort parents. In both the CHD and the comparison cohorts, mtDNAcns were significantly correlated between mother-child, father-child, and mother-father. mtDNAcns among people with European (mean = 52.0), African (53.0), and Asian haplogroups (53.5) were calculated and were significantly different for European and Asian haplogroups (p = 2.6E-3). Variant heteroplasmic fraction (HF) in blood correlated well with paired cardiovascular tissue HF (r = 0.975) and RNA VAF (r = 0.953), which suggests blood HF is a reasonable proxy for HF in heart tissue. We conclude that mtDNA mutations and mtDNAcns are unlikely to contribute significantly to CHD risk.
Keywords: congenital heart disease; genome sequencing; mitochondrial copy number; mitochondrial genome.
Copyright © 2022. Published by Elsevier Inc.
Conflict of interest statement
Declaration of interests The authors declare that they have no competing interests.
Figures

References
-
- van der Linde D., Konings E.E.M., Slager M.A., Witsenburg M., Helbing W.A., Takkenberg J.J.M., Roos-Hesselink J.W. Birth prevalence of congenital heart disease worldwide: a systematic review and meta-analysis. J. Am. Coll. Cardiol. 2011;58:2241–2247. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- UM1 HL098123/HL/NHLBI NIH HHS/United States
- T32 HL007572/HL/NHLBI NIH HHS/United States
- R01 HL080494/HL/NHLBI NIH HHS/United States
- R01 HL144776/HL/NHLBI NIH HHS/United States
- UM1 HL128711/HL/NHLBI NIH HHS/United States
- UM1 HL098147/HL/NHLBI NIH HHS/United States
- U01 HL098147/HL/NHLBI NIH HHS/United States
- UM1 HL172717/HL/NHLBI NIH HHS/United States
- U01 HL098163/HL/NHLBI NIH HHS/United States
- U01 HL128711/HL/NHLBI NIH HHS/United States
- UM1 HL098162/HL/NHLBI NIH HHS/United States
- P50 HD105351/HD/NICHD NIH HHS/United States
- U01 HL098188/HL/NHLBI NIH HHS/United States
- U01 HL098153/HL/NHLBI NIH HHS/United States
- U01 HL131003/HL/NHLBI NIH HHS/United States
- R03 HL150412/HL/NHLBI NIH HHS/United States
- UM1 HL128761/HL/NHLBI NIH HHS/United States
- R01 HL084553/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

