AdpA, a developmental regulator, promotes ε-poly-L-lysine biosynthesis in Streptomyces albulus
- PMID: 35397580
- PMCID: PMC8994273
- DOI: 10.1186/s12934-022-01785-6
AdpA, a developmental regulator, promotes ε-poly-L-lysine biosynthesis in Streptomyces albulus
Abstract
Background: AdpA is a global regulator of morphological differentiation and secondary metabolism in Streptomyces, but the regulatory roles of the Streptomyces AdpA family on the biosynthesis of the natural product ε-poly-L-lysine (ε-PL) remain unidentified, and few studies have focused on increasing the production of ε-PL by manipulating transcription factors in Streptomyces.
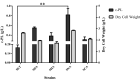
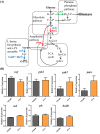
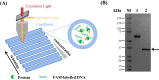
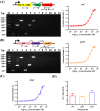
Results: In this study, we revealed the regulatory roles of different AdpA homologs in ε-PL biosynthesis and morphological differentiation and effectively promoted ε-PL production and sporulation in Streptomyces albulus NK660 by heterologously expressing adpA from S. neyagawaensis NRRLB-3092 (adpASn). First, we identified a novel AdpA homolog named AdpASa in S. albulus NK660 and characterized its function as an activator of ε-PL biosynthesis and morphological differentiation. Subsequently, four heterologous AdpA homologs were selected to investigate their phylogenetic relationships and regulatory roles in S. albulus, and AdpASn was demonstrated to have the strongest ability to promote both ε-PL production and sporulation among these five AdpA proteins. The ε-PL yield of S. albulus heterologously expressing adpASn was approximately 3.6-fold higher than that of the control strain. Finally, we clarified the mechanism of AdpASn in enhancing ε-PL biosynthesis and its effect on ε-PL polymerization degree using real-time quantitative PCR, microscale thermophoresis and MALDI-TOF-MS. AdpASn was purified, and its seven direct targets, zwf, tal, pyk2, pta, ack, pepc and a transketolase gene (DC74_2409), were identified, suggesting that AdpASn may cause the redistribution of metabolic flux in central metabolism pathways, which subsequently provides more carbon skeletons and ATP for ε-PL biosynthesis in S. albulus.
Conclusions: Here, we characterized the positive regulatory roles of Streptomyces AdpA homologs in ε-PL biosynthesis and their effects on morphological differentiation and reported for the first time that AdpASn promotes ε-PL biosynthesis by affecting the transcription of its target genes in central metabolism pathways. These findings supply valuable insights into the regulatory roles of the Streptomyces AdpA family on ε-PL biosynthesis and morphological differentiation and suggest that AdpASn may be an effective global regulator for enhanced production of ε-PL and other valuable secondary metabolites in Streptomyces.
Keywords: AdpA; Morphological differentiation; Polymerization degree; Streptomyces albulus; ε-Poly-L-lysine.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures





Similar articles
-
Enhanced ε-Poly-L-Lysine Production in Streptomyces albulus through Multi-Omics-Guided Metabolic Engineering.Biomolecules. 2024 Jun 25;14(7):752. doi: 10.3390/biom14070752. Biomolecules. 2024. PMID: 39062465 Free PMC article.
-
Cloning of ε-poly-L-lysine (ε-PL) synthetase gene from a newly isolated ε-PL-producing Streptomyces albulus NK660 and its heterologous expression in Streptomyces lividans.Microb Biotechnol. 2014 Mar;7(2):155-64. doi: 10.1111/1751-7915.12108. Epub 2014 Jan 14. Microb Biotechnol. 2014. PMID: 24423427 Free PMC article.
-
Effects of Chromosomal Integration of the Vitreoscilla Hemoglobin Gene (vgb) and S-Adenosylmethionine Synthetase Gene (metK) on ε-Poly-L-Lysine Synthesis in Streptomyces albulus NK660.Appl Biochem Biotechnol. 2016 Apr;178(7):1445-57. doi: 10.1007/s12010-015-1958-7. Epub 2016 Jan 9. Appl Biochem Biotechnol. 2016. PMID: 26749294
-
epsilon-Poly-L-lysine: microbial production, biodegradation and application potential.Appl Microbiol Biotechnol. 2003 Jul;62(1):21-6. doi: 10.1007/s00253-003-1312-9. Epub 2003 May 1. Appl Microbiol Biotechnol. 2003. PMID: 12728342 Review.
-
Advances in ε-Poly-Lysine Biosynthesis, Selection of High-Yielding Strains and Regulatory Mechanisms.Biotechnol J. 2025 Sep;20(9):e70111. doi: 10.1002/biot.70111. Biotechnol J. 2025. PMID: 40891134 Review.
Cited by
-
Genomic and Metabolomic Analyses of Streptomyces albulus with Enhanced ε-Poly-l-lysine Production Through Adaptive Laboratory Evolution.Microorganisms. 2025 Jan 13;13(1):149. doi: 10.3390/microorganisms13010149. Microorganisms. 2025. PMID: 39858917 Free PMC article.
-
The Pleiotropic Regulator AdpA Regulates the Removal of Excessive Sulfane Sulfur in Streptomyces coelicolor.Antioxidants (Basel). 2023 Jan 29;12(2):312. doi: 10.3390/antiox12020312. Antioxidants (Basel). 2023. PMID: 36829871 Free PMC article.
-
Engineering Streptomyces albulus to enhance ε-poly-L-lysine production by introducing a polyphosphate kinase-mediated ATP regeneration system.Microb Cell Fact. 2023 Mar 14;22(1):51. doi: 10.1186/s12934-023-02057-7. Microb Cell Fact. 2023. PMID: 36918890 Free PMC article.
-
Enhanced ε-Poly-L-Lysine Production in Streptomyces albulus through Multi-Omics-Guided Metabolic Engineering.Biomolecules. 2024 Jun 25;14(7):752. doi: 10.3390/biom14070752. Biomolecules. 2024. PMID: 39062465 Free PMC article.
-
A novel self-assembled nanocarrier-mediated dsRNA fungicide for broad-spectrum management of Rhizoctonia solani.Mater Today Bio. 2025 Jul 16;34:102099. doi: 10.1016/j.mtbio.2025.102099. eCollection 2025 Oct. Mater Today Bio. 2025. PMID: 40735700 Free PMC article.
References
-
- Flärdh K, Buttner MJ. Streptomyces morphogenetics: dissecting differentiation in a filamentous bacterium. Nat Rev Microbiol. 2009;7:36–49. - PubMed
-
- Bush MJ, Tschowri N, Schlimpert S, Flärdh K, Buttner MJ. c-di-GMP signalling and the regulation of developmental transitions in streptomycetes. Nat Rev Microbiol. 2015;13:749–760. - PubMed
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

