The dimerization mechanism of the N-terminal domain of spider silk proteins is conserved despite extensive sequence divergence
- PMID: 35398358
- PMCID: PMC9097459
- DOI: 10.1016/j.jbc.2022.101913
The dimerization mechanism of the N-terminal domain of spider silk proteins is conserved despite extensive sequence divergence
Abstract
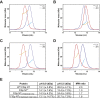
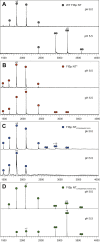
The N-terminal (NT) domain of spider silk proteins (spidroins) is crucial for their storage at high concentrations and also regulates silk assembly. NTs from the major ampullate spidroin (MaSp) and the minor ampullate spidroin are monomeric at neutral pH and confer solubility to spidroins, whereas at lower pH, they dimerize to interconnect spidroins in a fiber. This dimerization is known to result from modulation of electrostatic interactions by protonation of well-conserved glutamates, although it is undetermined if this mechanism applies to other spidroin types as well. Here, we determine the solution and crystal structures of the flagelliform spidroin NT, which shares only 35% identity with MaSp NT, and investigate the mechanisms of its dimerization. We show that flagelliform spidroin NT is structurally similar to MaSp NT and that the electrostatic intermolecular interaction between Asp 40 and Lys 65 residues is conserved. However, the protonation events involve a different set of residues than in MaSp, indicating that an overall mechanism of pH-dependent dimerization is conserved but can be mediated by different pathways in different silk types.
Keywords: NMR structure; X-ray structure; dimerization; protein domain; silk assembly; spidroin.
Copyright © 2022 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures





References
-
- Peakall D.B. Synthesis of silk, mechanism and location. Am. Zool. 1969;9:71–79.
-
- Gosline J.M., Guerette P.A., Ortlepp C.S., Savage K.N. The mechanical design of spider silks: From fibroin sequence to mechanical function. J. Exp. Biol. 1999;202:3295–3303. - PubMed
-
- Casem M.L., Tran L.P., Moore A.M. Ultrastructure of the major ampullate gland of the black widow spider, Latrodectus hesperus. Tissue Cell. 2002;34:427–436. - PubMed
-
- Giesa T., Perry C.C., Buehler M.J. Secondary structure transition and critical stress for a model of spider silk assembly. Biomacromolecules. 2016;17:427–436. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

