PD-1 Blockade in Solid Tumors with Defects in Polymerase Epsilon
- PMID: 35398880
- PMCID: PMC9167784
- DOI: 10.1158/2159-8290.CD-21-0521
PD-1 Blockade in Solid Tumors with Defects in Polymerase Epsilon
Abstract
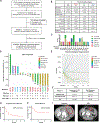
Missense mutations in the polymerase epsilon (POLE) gene have been reported to generate proofreading defects resulting in an ultramutated genome and to sensitize tumors to checkpoint blockade immunotherapy. However, many POLE-mutated tumors do not respond to such treatment. To better understand the link between POLE mutation variants and response to immunotherapy, we prospectively assessed the efficacy of nivolumab in a multicenter clinical trial in patients bearing advanced mismatch repair-proficient POLE-mutated solid tumors. We found that only tumors harboring selective POLE pathogenic mutations in the DNA binding or catalytic site of the exonuclease domain presented high mutational burden with a specific single-base substitution signature, high T-cell infiltrates, and a high response rate to anti-PD-1 monotherapy. This study illustrates how specific DNA repair defects sensitize to immunotherapy. POLE proofreading deficiency represents a novel agnostic biomarker for response to PD-1 checkpoint blockade therapy.
Significance: POLE proofreading deficiency leads to high tumor mutational burden with high tumor-infiltrating lymphocytes and predicts anti-PD-1 efficacy in mismatch repair-proficient tumors. Conversely, tumors harboring POLE mutations not affecting proofreading derived no benefit from PD-1 blockade. POLE proofreading deficiency is a new tissue-agnostic biomarker for cancer immunotherapy. This article is highlighted in the In This Issue feature, p. 1397.
©2022 American Association for Cancer Research.
Figures



References
-
- Rayner E, van Gool IC, Palles C, Kearsey SE, Bosse T, Tomlinson I, et al. A panoply of errors: polymerase proofreading domain mutations in cancer. Nat Rev Cancer. 2016;16:71–81. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

