Honeysuckle (Lonicera japonica) and Huangqi (Astragalus membranaceus) Suppress SARS-CoV-2 Entry and COVID-19 Related Cytokine Storm in Vitro
- PMID: 35401158
- PMCID: PMC8990830
- DOI: 10.3389/fphar.2021.765553
Honeysuckle (Lonicera japonica) and Huangqi (Astragalus membranaceus) Suppress SARS-CoV-2 Entry and COVID-19 Related Cytokine Storm in Vitro
Abstract
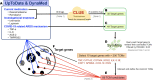
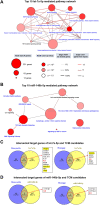
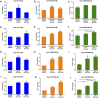
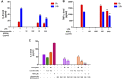
COVID-19 is threatening human health worldwide but no effective treatment currently exists for this disease. Current therapeutic strategies focus on the inhibition of viral replication or using anti-inflammatory/immunomodulatory compounds to improve host immunity, but not both. Traditional Chinese medicine (TCM) compounds could be promising candidates due to their safety and minimal toxicity. In this study, we have developed a novel in silico bioinformatics workflow that integrates multiple databases to predict the use of honeysuckle (Lonicera japonica) and Huangqi (Astragalus membranaceus) as potential anti-SARS-CoV-2 agents. Using extracts from honeysuckle and Huangqi, these two herbs upregulated a group of microRNAs including let-7a, miR-148b, and miR-146a, which are critical to reduce the pathogenesis of SARS-CoV-2. Moreover, these herbs suppressed pro-inflammatory cytokines including IL-6 or TNF-α, which were both identified in the cytokine storm of acute respiratory distress syndrome, a major cause of COVID-19 death. Furthermore, both herbs partially inhibited the fusion of SARS-CoV-2 spike protein-transfected BHK-21 cells with the human lung cancer cell line Calu-3 that was expressing ACE2 receptors. These herbs inhibited SARS-CoV-2 Mpro activity, thereby alleviating viral entry as well as replication. In conclusion, our findings demonstrate that honeysuckle and Huangqi have the potential to be used as an inhibitor of SARS-CoV-2 virus entry that warrants further in vivo analysis and functional assessment of miRNAs to confirm their clinical importance. This fast-screening platform can also be applied to other drug discovery studies for other infectious diseases.
Keywords: COVID-19; Huangqi; SARS-CoV-2; honeysuckle; let-7a; miR-148b; microRNA; mir-146a.
Copyright © 2022 Yeh, Doan, Huang, Chu, Shi, Lee, Wu, Lin, Chiang, Liu, Chuang, Ping, Liu and Huang.
Conflict of interest statement
Author C-TW was employed by the company Phalanx Biotech Group. Author S-TC was employed by Chuang Song Zong Pharmaceutical Co., Ltd. Ligang Plant. Author Z-YH received a scholarship from ASUS Intelligent Cloud Services. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Honeysuckle Aqueous Extracts Induced let-7a Suppress EV71 Replication and Pathogenesis In Vitro and In Vivo and Is Predicted to Inhibit SARS-CoV-2.Viruses. 2021 Feb 16;13(2):308. doi: 10.3390/v13020308. Viruses. 2021. PMID: 33669264 Free PMC article.
-
Repurposing Astragalus Polysaccharide PG2 for Inhibiting ACE2 and SARS-CoV-2 Spike Syncytial Formation and Anti-Inflammatory Effects.Viruses. 2023 Feb 27;15(3):641. doi: 10.3390/v15030641. Viruses. 2023. PMID: 36992350 Free PMC article.
-
Honeysuckle aqueous extract and induced let-7a suppress dengue virus type 2 replication and pathogenesis.J Ethnopharmacol. 2017 Feb 23;198:109-121. doi: 10.1016/j.jep.2016.12.049. Epub 2017 Jan 2. J Ethnopharmacol. 2017. PMID: 28052239
-
Tocilizumab in SARS-CoV-2 Patients with the Syndrome of Cytokine Storm: A Narrative Review.Rev Recent Clin Trials. 2021;16(2):138-145. doi: 10.2174/1574887115666200917110954. Rev Recent Clin Trials. 2021. PMID: 32940187 Review.
-
Potential Role of Gut Microbiota in Traditional Chinese Medicine against COVID-19.Am J Chin Med. 2021;49(4):785-803. doi: 10.1142/S0192415X21500373. Epub 2021 Apr 9. Am J Chin Med. 2021. PMID: 33853498 Review.
Cited by
-
Network analysis-based strategy to investigate the protective effect of cepharanthine on rat acute respiratory distress syndrome.Front Pharmacol. 2022 Oct 26;13:1054339. doi: 10.3389/fphar.2022.1054339. eCollection 2022. Front Pharmacol. 2022. PMID: 36386130 Free PMC article.
-
Practices, Knowledge, and Attitudes of Chinese University Students Toward Traditional Chinese Herbal Medicine for the Control of COVID-19.Infect Drug Resist. 2022 Nov 30;15:6951-6962. doi: 10.2147/IDR.S387292. eCollection 2022. Infect Drug Resist. 2022. PMID: 36474905 Free PMC article.
-
A review on NLRP3 inflammasome modulation by animal venom proteins/peptides: mechanisms and therapeutic insights.Inflammopharmacology. 2025 Mar;33(3):1013-1031. doi: 10.1007/s10787-025-01656-7. Epub 2025 Feb 11. Inflammopharmacology. 2025. PMID: 39934538 Review.
-
Network pharmacology combined with metabolomics to explore the mechanism for Lonicerae Japonicae flos against respiratory syncytial virus.BMC Complement Med Ther. 2023 Dec 12;23(1):449. doi: 10.1186/s12906-023-04286-0. BMC Complement Med Ther. 2023. PMID: 38087272 Free PMC article.
-
Analyzing the Material Basis of Anti-RSV Efficacy of Lonicerae japonicae Flos Based on the PK-PD Model.Molecules. 2023 Sep 5;28(18):6437. doi: 10.3390/molecules28186437. Molecules. 2023. PMID: 37764214 Free PMC article.
References
-
- Aleem A., Akbar Samad A. B., Slenker A. K. (2021). Emerging Variants of SARS-CoV-2 and Novel Therapeutics against Coronavirus (COVID-19). Treasure Island (FL): StatPearls.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

