Uncovering Essential Tremor Genetics: The Promise of Long-Read Sequencing
- PMID: 35401394
- PMCID: PMC8983820
- DOI: 10.3389/fneur.2022.821189
Uncovering Essential Tremor Genetics: The Promise of Long-Read Sequencing
Abstract
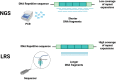
Long-read sequencing (LRS) technologies have been recently introduced to overcome intrinsic limitations of widely-used next-generation sequencing (NGS) technologies, namely the sequencing limited to short-read fragments (150-300 base pairs). Since its introduction, LRS has permitted many successes in unraveling hidden mutational mechanisms. One area in clinical neurology in need of rethinking as it applies to genetic mechanisms is essential tremor (ET). This disorder, among the most common in neurology, is a syndrome often exhibiting an autosomal dominant pattern of inheritance whose large phenotypic spectrum suggest a multitude of genetic etiologies. Exome sequencing has revealed the genetic etiology only in rare ET families (FUS, SORT1, SCN4A, NOS3, KCNS2, HAPLN4/BRAL2, and USP46). We hypothesize that a reason for this shortcoming may be non-classical genetic mechanism(s) underpinning ET, among them trinucleotide, tetranucleotide, or pentanucleotide repeat disorders. In support of this hypothesis, trinucleotide (e.g., GGC repeats in NOTCH2NLC) and pentanucleotide repeat disorders (e.g., ATTTC repeats in STARD7) have been revealed as pathogenic in patients with a past history of what has come to be referred to as "ET plus," bilateral hand tremor associated with epilepsy and/or leukoencephalopathy. A systematic review of LRS in neurodegenerative disorders showed that 10 of the 22 (45%) genetic etiologies ascertained by LRS include tremor in their phenotypic spectrum, suggesting that future clinical applications of LRS for tremor disorders may uncover genetic subtypes of familial ET that have eluded NGS, particularly those with associated leukoencephalopathy or family history of epilepsy. LRS provides a pathway for potentially uncovering novel genes and genetic mechanisms, helping narrow the large proportion of "idiopathic" ET.
Keywords: genomics; long-read sequencing; movement disorders; tremor; whole-genome sequencing.
Copyright © 2022 Marsili, Duque, Bode, Kauffman and Espay.
Conflict of interest statement
LM has received honoraria from the International Association of Parkinsonism and Related Disorders (IAPRD) Society for social media and web support. MK is an employee of the CONICET and has received grant support from the Ministry of Science and Technology of Argentina and the Ministry of Health of Buenos Aires. AE has received grant support from the NIH and the Michael J Fox Foundation; personal compensation as a consult-ant/scientific advisory board member for Abbvie, Neuroderm, Neurocrine, Amneal, Adamas, Acadia, Acorda, Kyowa Kirin, Sunovion, Lundbeck, and USWorldMeds; publishing royalties from Lippincott Williams & Wilkins, Cambridge University Press, and Springer; and honoraria from USWorldMeds, Acadia, and Sunovion. He is cofounder of REGAIN Therapeutics, owner of a provisional patent on compositions and methods for treatment and/or prophylaxis of proteinopathies. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

