Functional and Phylogenetic Characterization of Bacteria in Bovine Rumen Using Fractionation of Ruminal Fluid
- PMID: 35401437
- PMCID: PMC8992543
- DOI: 10.3389/fmicb.2022.813002
Functional and Phylogenetic Characterization of Bacteria in Bovine Rumen Using Fractionation of Ruminal Fluid
Abstract
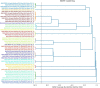
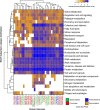
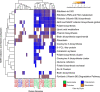
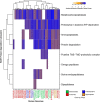
Cattle productivity depends on our ability to fully understand and manipulate the fermentation process of plant material that occurs in the bovine rumen, which ultimately leads to the improvement of animal health and increased productivity with a reduction in environmental impact. An essential step in this direction is the phylogenetic and functional characterization of the microbial species composing the ruminal microbiota. To address this challenge, we separated a ruminal fluid sample by size and density using a sucrose density gradient. We used the full sample and the smallest fraction (5%), allowing the enrichment of bacteria, to assemble metagenome-assembled genomes (MAGs). We obtained a total of 16 bacterial genomes, 15 of these enriched in the smallest fraction of the gradient. According to the recently proposed Genome Taxonomy Database (GTDB) taxonomy, these MAGs belong to Bacteroidota, Firmicutes_A, Firmicutes, Proteobacteria, and Spirochaetota phyla. Fifteen MAGs were novel at the species level and four at the genus level. The functional characterization of these MAGs suggests differences from what is currently known from the genomic potential of well-characterized members from this complex environment. Species of the phyla Bacteroidota and Spirochaetota show the potential for hydrolysis of complex polysaccharides in the plant cell wall and toward the production of B-complex vitamins and protein degradation in the rumen. Conversely, the MAGs belonging to Firmicutes and Alphaproteobacteria showed a reduction in several metabolic pathways; however, they have genes for lactate fermentation and the presence of hydrolases and esterases related to chitin degradation. Our results demonstrate that the separation of the rumen microbial community by size and density reduced the complexity of the ruminal fluid sample and enriched some poorly characterized ruminal bacteria allowing exploration of their genomic potential and their functional role in the rumen ecosystem.
Keywords: MAGs; fractionation; new species; ruminal fluid; small-sized bacteria.
Copyright © 2022 Hernández, Chaib De Mares, Jimenez, Reyes and Caro-Quintero.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
Recovery of metagenome-assembled genomes from the rumen and fecal microbiomes of Bos indicus beef cattle.Sci Data. 2024 Dec 18;11(1):1385. doi: 10.1038/s41597-024-04271-3. Sci Data. 2024. PMID: 39695297 Free PMC article.
-
Metagenomic Analysis of in Vitro Ruminal Fermentation Reveals the Role of the Copresent Microbiome in Plant Biomass Degradation.J Agric Food Chem. 2022 Sep 28;70(38):12095-12106. doi: 10.1021/acs.jafc.2c03522. Epub 2022 Sep 19. J Agric Food Chem. 2022. PMID: 36121066
-
The bovine epimural microbiota displays compositional and structural heterogeneity across different ruminal locations.J Dairy Sci. 2020 Apr;103(4):3636-3647. doi: 10.3168/jds.2019-17649. Epub 2020 Feb 11. J Dairy Sci. 2020. PMID: 32057427
-
Ruminal acidosis, bacterial changes, and lipopolysaccharides.J Anim Sci. 2020 Aug 1;98(8):skaa248. doi: 10.1093/jas/skaa248. J Anim Sci. 2020. PMID: 32761212 Free PMC article. Review.
-
A Genomic Perspective Across Earth's Microbiomes Reveals That Genome Size in Archaea and Bacteria Is Linked to Ecosystem Type and Trophic Strategy.Front Microbiol. 2022 Jan 5;12:761869. doi: 10.3389/fmicb.2021.761869. eCollection 2021. Front Microbiol. 2022. PMID: 35069467 Free PMC article. Review.
Cited by
-
Effect of differences in residual feed intake on gastrointestinal microbiota of Dexin fine-wool meat sheep.Front Microbiol. 2024 Dec 20;15:1482017. doi: 10.3389/fmicb.2024.1482017. eCollection 2024. Front Microbiol. 2024. PMID: 39760076 Free PMC article.
-
Rumen-Degradable Starch Improves Rumen Fermentation, Function, and Growth Performance by Altering Bacteria and Its Metabolome in Sheep Fed Alfalfa Hay or Silage.Animals (Basel). 2024 Dec 26;15(1):34. doi: 10.3390/ani15010034. Animals (Basel). 2024. PMID: 39794977 Free PMC article.
-
Enhancing Biodiversity-Function Relationships in Field Retting: Towards Key Microbial Indicators for Retting Control.Environ Microbiol Rep. 2025 Jun;17(3):e70102. doi: 10.1111/1758-2229.70102. Environ Microbiol Rep. 2025. PMID: 40531656 Free PMC article.
-
Effects of residual black wolfberry fruit on growth performance, rumen fermentation parameters, microflora and economic benefits of fattening sheep.Front Vet Sci. 2025 Jan 10;11:1528126. doi: 10.3389/fvets.2024.1528126. eCollection 2024. Front Vet Sci. 2025. PMID: 39867601 Free PMC article.
-
Comparison of ruminal microbiota, IL-1β gene variation, and tick incidence between Holstein × Gyr and Holstein heifers in grazing system.Front Microbiol. 2024 Feb 26;15:1132151. doi: 10.3389/fmicb.2024.1132151. eCollection 2024. Front Microbiol. 2024. PMID: 38468851 Free PMC article.
References
-
- Alneberg J., Bjarnason B. S., de Bruijn I., Schirmer M., Quick J., Ijaz U. Z., et al. (2013). CONCOCT: Clustering cONtigs on COverage and ComposiTion. ArXiv13124038 Q-Bio. Available online at: http://arxiv.org/abs/1312.4038 (accessed June 9, 2021).
-
- Andrews S. (2010). FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed April 2020).
LinkOut - more resources
Full Text Sources

