Persistent Overactive Cytotoxic Immune Response in a Spanish Cohort of Individuals With Long-COVID: Identification of Diagnostic Biomarkers
- PMID: 35401523
- PMCID: PMC8990790
- DOI: 10.3389/fimmu.2022.848886
Persistent Overactive Cytotoxic Immune Response in a Spanish Cohort of Individuals With Long-COVID: Identification of Diagnostic Biomarkers
Abstract
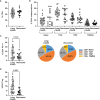
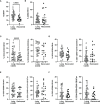
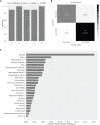
Long-COVID is a new emerging syndrome worldwide that is characterized by the persistence of unresolved signs and symptoms of COVID-19 more than 4 weeks after the infection and even after more than 12 weeks. The underlying mechanisms for Long-COVID are still undefined, but a sustained inflammatory response caused by the persistence of SARS-CoV-2 in organ and tissue sanctuaries or resemblance with an autoimmune disease are within the most considered hypotheses. In this study, we analyzed the usefulness of several demographic, clinical, and immunological parameters as diagnostic biomarkers of Long-COVID in one cohort of Spanish individuals who presented signs and symptoms of this syndrome after 49 weeks post-infection, in comparison with individuals who recovered completely in the first 12 weeks after the infection. We determined that individuals with Long-COVID showed significantly increased levels of functional memory cells with high antiviral cytotoxic activity such as CD8+ TEMRA cells, CD8±TCRγδ+ cells, and NK cells with CD56+CD57+NKG2C+ phenotype. The persistence of these long-lasting cytotoxic populations was supported by enhanced levels of CD4+ Tregs and the expression of the exhaustion marker PD-1 on the surface of CD3+ T lymphocytes. With the use of these immune parameters and significant clinical features such as lethargy, pleuritic chest pain, and dermatological injuries, as well as demographic factors such as female gender and O+ blood type, a Random Forest algorithm predicted the assignment of the participants in the Long-COVID group with 100% accuracy. The definition of the most accurate diagnostic biomarkers could be helpful to detect the development of Long-COVID and to improve the clinical management of these patients.
Keywords: CD8+ T cells; Long-COVID; NK cells; Random Forest algorithm; cytotoxic immune response; immune exhaustion.
Copyright © 2022 Galán, Vigón, Fuertes, Murciano-Antón, Casado-Fernández, Domínguez-Mateos, Mateos, Ramos-Martín, Planelles, Torres, Rodríguez-Mora, López-Huertas and Coiras.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- WHO . Coronavirus (COVID-19) Dashboard. Available at: https://covid19.who.int/.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

